5JZH
 
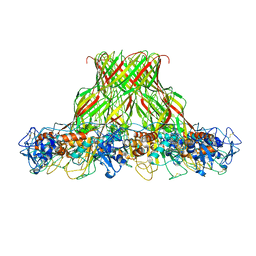 | | Cryo-EM structure of aerolysin prepore | | Descriptor: | Aerolysin | | Authors: | Iacovache, I, Zuber, B. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-13 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of aerolysin variants reveals a novel protein fold and the pore-formation process.
Nat Commun, 7, 2016
|
|
5JZW
 
 | |
5JZT
 
 | |
7Q9Y
 
 | |
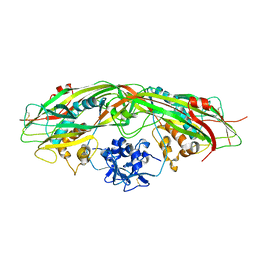
3G4O
 
 | |
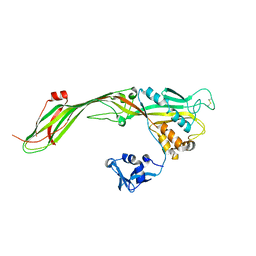
3G4N
 
 | |
3C0N
 
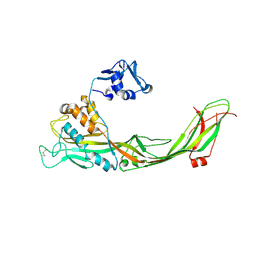 | | Crystal structure of the proaerolysin mutant Y221G at 2.2 A | | Descriptor: | Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3C0O
 
 | | Crystal structure of the proaerolysin mutant Y221G complexed with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, ACETATE ION, Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3C0M
 
 | | Crystal structure of the proaerolysin mutant Y221G | | Descriptor: | Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
