8DQV
 
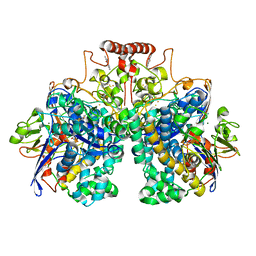 | | The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-07-20 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.52 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUR
 
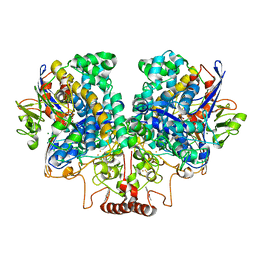 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
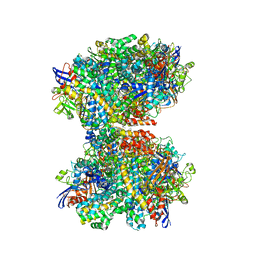 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-26 | | Release date: | 2023-01-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | Authors: | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-04 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
6E4V
 
 | |
6OFS
 
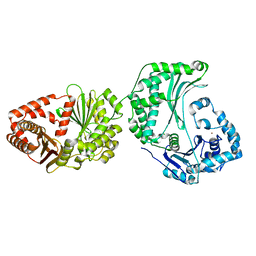 | |
6OFT
 
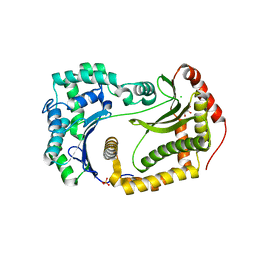 | |
6OFR
 
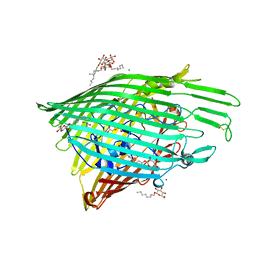 | |
4ZHO
 
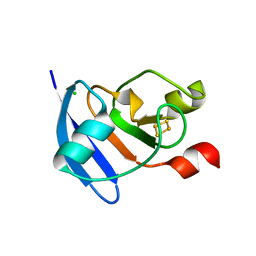 | | The crystal structure of Arabidopsis ferredoxin 2 with 2Fe-2S cluster | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZHP
 
 | | The crystal structure of Potato ferredoxin I with 2Fe-2S cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Potato Ferredoxin I | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZGV
 
 | | The Crystal Structure of the Ferredoxin Receptor FusA from Pectobacterium atrosepticum SCRI1043 | | Descriptor: | Ferredoxin receptor, LAURYL DIMETHYLAMINE-N-OXIDE, octyl beta-D-glucopyranoside | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-24 | | Release date: | 2016-08-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
6UW1
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UVX
 
 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Apo state | | Descriptor: | CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW5
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, GDP and Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW7
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, Dehydro-F420-0 bound form | | Descriptor: | 2-[oxidanyl-[(2~{R},3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-[2,4,8-tris(oxidanylidene)-1,9-dihydropyrimido[4,5-b]quinolin-10-yl]pentoxy]phosphoryl]oxyprop-2-enoic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Izore, T, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6UW3
 
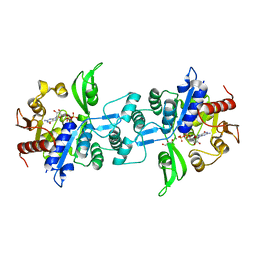 | | The crystal structure of FbiA from Mycobacterium Smegmatis, GDP Bound form | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6B05
 
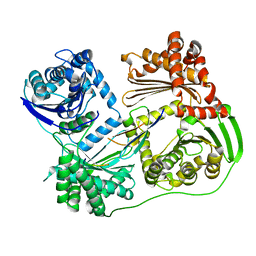 | |
6B03
 
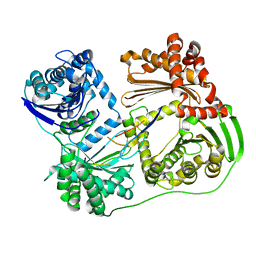 | |
6BPM
 
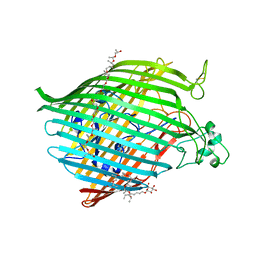 | | The crystal structure of the Ferric-Catecholate import receptor Fiu from K12 E. coli: Closed form (C21) | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, Catecholate siderophore receptor Fiu, octyl beta-D-glucopyranoside | | Authors: | Grinter, R. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of the bacterial iron-catecholate transporter Fiu suggests that it imports substrates via a two-step mechanism.
J.Biol.Chem., 2019
|
|
6BRS
 
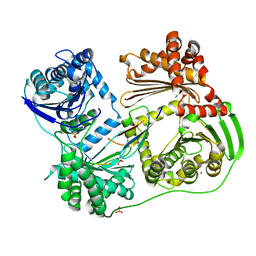 | |
6BPN
 
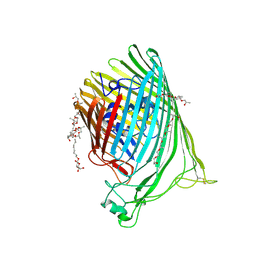 | | The crystal structure of the Ferric-Catecholate import receptor Fiu from E. coli K12: Open form (C2221) | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Grinter, R. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the bacterial iron-catecholate transporter Fiu suggests that it imports substrates via a two-step mechanism.
J.Biol.Chem., 2019
|
|
6BPO
 
 | |
4N58
 
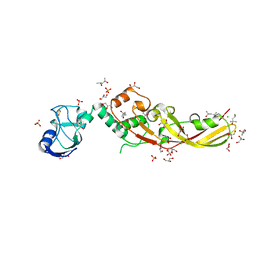 | | Crystal Structure of Pectocin M2 at 1.86 Angstroms | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Grinter, R, Roszak, A.W, Zeth, K, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
4QKO
 
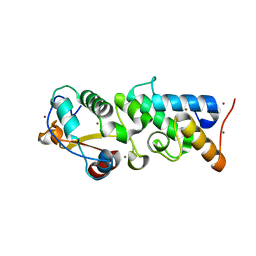 | | The Crystal Structure of the Pyocin S2 Nuclease Domain, Immunity Protein Complex at 1.8 Angstroms | | Descriptor: | BROMIDE ION, MAGNESIUM ION, Pyocin-S2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, C.J, Walker, D. | | Deposit date: | 2014-06-07 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into pyocin S2
To be Published
|
|
6V81
 
 | | The crystal structure of the outer-membrane transporter YncD | | Descriptor: | CALCIUM ION, Probable TonB-dependent receptor YncD, octyl beta-D-glucopyranoside | | Authors: | Grinter, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | The crystal structure of the TonB-dependent transporter YncD reveals a positively charged substrate-binding site.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
