1FW8
 
 | | CIRCULARLY PERMUTED PHOSPHOGLYCERATE KINASE FROM YEAST: PGK P72 | | Descriptor: | GLYCEROL, Phosphoglycerate kinase | | Authors: | Tougard, P, Bizebard, T, Ritco-Vonsovici, M, Minard, P, Desmadril, M. | | Deposit date: | 2000-09-22 | | Release date: | 2001-03-22 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a circularly permuted phosphoglycerate kinase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1W8F
 
 | | PSEUDOMONAS AERUGINOSA LECTIN II (PA-IIL)COMPLEXED WITH LACTO-N-NEO- FUCOPENTAOSE V(LNPFV) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
1W8H
 
 | | structure of pseudomonas aeruginosa lectin II (PA-IIL)complexed with lewisA trisaccharide | | Descriptor: | CALCIUM ION, GLYCEROL, PSEUDOMONAS AERUGINOSA LECTIN II, ... | | Authors: | Perret, S, Sabin, C, Dumon, C, Budova, M, Gautier, C, Galanina, O, Ilia, S, Bovin, N, Nicaise, M, Desmadril, M, Gilboa-Garber, N, Wimmerova, M, Mitchell, E.P, Imberty, A. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Interaction between Human Milk Oligosaccharides and the Bacterial Lectin Pa-Iil of Pseudomonas Aeruginosa.
Biochem.J., 389, 2005
|
|
2CBM
 
 | | Crystal structure of the apo-form of a neocarzinostatin mutant evolved to bind testosterone. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NEOCARZINOSTATIN | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBQ
 
 | | Crystal structure of the neocarzinostatin 1Tes15 mutant bound to testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, SULFATE ION, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBO
 
 | | Crystal structure of the neocarzinostatin 3Tes24 mutant bound to testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, SULFATE ION, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
2CBT
 
 | | Crystal structure of the neocarzinostatin 4Tes1 mutant bound testosterone hemisuccinate. | | Descriptor: | NEOCARZINOSTATIN, TESTOSTERONE HEMISUCCINATE | | Authors: | Drevelle, A, Graille, M, Heyd, B, Sorel, I, Ulryck, N, Pecorari, F, Desmadril, M, Van Tilbeurgh, H, Minard, P. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of in Vitro Evolved Binding Sites on Neocarzinostatin Scaffold Reveal Unanticipated Evolutionary Pathways.
J.Mol.Biol., 358, 2006
|
|
4JW3
 
 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | Alpha-helical artificial proteins, Neocarzinostatin | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
4JW2
 
 | | Selection of specific protein binders for pre-defined targets from an optimized library of artificial helicoidal repeat proteins (alphaRep) | | Descriptor: | 1,2-ETHANEDIOL, A3 artificial protein, bA3-2: binder of A3 protein | | Authors: | Guellouz, A, Valerio-Lepiniec, M, Urvoas, A, Chevrel, A, Graille, M, Fourati-Kammoun, Z, Desmadril, M, van Tilbeurgh, H, Minard, P. | | Deposit date: | 2013-03-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selection of Specific Protein Binders for Pre-Defined Targets from an Optimized Library of Artificial Helicoidal Repeat Proteins (alphaRep).
Plos One, 8, 2013
|
|
3LTM
 
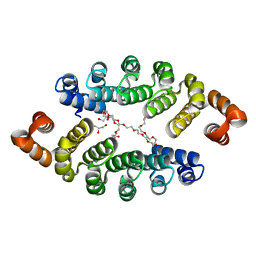 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
3LTJ
 
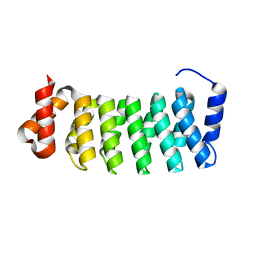 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | AlphaRep-4 | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
6RCY
 
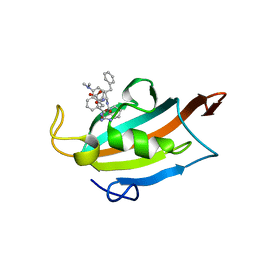 | |
6HWP
 
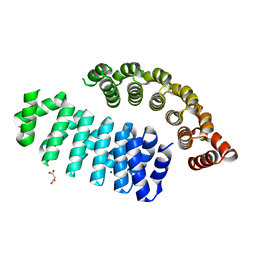 | |
4XL5
 
 | | X-ray structure of bGFP-A / EGFP complex | | Descriptor: | Green fluorescent protein, bGFP-A | | Authors: | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
4XVP
 
 | | X-ray structure of bGFP-C / EGFP complex | | Descriptor: | BGFP-C, Green fluorescent protein | | Authors: | Chevrel, A, Urvoas, A, Li de la Sierra-Gallay, I, Van Tilbeurgh, H, Minard, P, Valerio-Lepiniec, M. | | Deposit date: | 2015-01-27 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Specific GFP-binding artificial proteins ( alpha Rep): a new tool for in vitro to live cell applications.
Biosci.Rep., 35, 2015
|
|
6FT5
 
 | | Structure of A3_A3, an artificial bi-domain protein based on two identical alphaRep A3 domains | | Descriptor: | GLYCEROL, SULFATE ION, alphaRep A3_A3 | | Authors: | Li de la Sierra-Gallay, I, Leger, C, Di Meo, T. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Ligand-induced conformational switch in an artificial bidomain protein scaffold.
Sci Rep, 9, 2019
|
|
6FSQ
 
 | |
5MTE
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T in complex with actinonin - crystal form II | | Descriptor: | ACTINONIN, NICKEL (II) ION, Putative uncharacterized protein orf60T, ... | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptide deformylases from Vibrio parahaemolyticus phage and bacteria display similar deformylase activity and inhibitor binding clefts.
Biochim. Biophys. Acta, 1866, 2018
|
|
2JDH
 
 | | Lectin PA-IIL of P.aeruginosa complexed with disaccharide derivative | | Descriptor: | 2H-1,2,3-TRIAZOL-4-YLMETHANOL, CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, ... | | Authors: | Marotte, K, Sabin, C, Preville, C, Pymbock, M, Deguise, I, Wimmerova, M, Mitchell, E.P, Imberty, A, Roy, R. | | Deposit date: | 2007-01-09 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-Ray Structures and Thermodynamics of the Interaction of Pa-Iil from Pseudomonas Aeruginosa with Disaccharide Derivatives.
Chemmedchem, 2, 2007
|
|
2VUC
 
 | | PA-IIL lectin from Pseudomonas aeruginosa complexed with Fucose- derived glycomimetics | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Beha, S, Marotte, K, Sabin, C, Mitchell, E.P, Imberty, A, Roy, R. | | Deposit date: | 2008-05-22 | | Release date: | 2009-07-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fucose-Derived Glycomimetics as High Affinity Ligands for Bacterial Lectin Pa-Iil from Pseudomonas Aeruginosa
To be Published
|
|
4JE8
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with Met-Ala-Ser | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JE7
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with actinonin | | Descriptor: | ACTINONIN, Peptide deformylase 1A, chloroplastic/mitochondrial, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JE6
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3M6Q
 
 | |
3M6R
 
 | |
