1X82
 
 | | CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE FROM PYROCOCCUS FURIOSUS WITH BOUND 5-phospho-D-arabinonate | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-17 | | Release date: | 2004-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X7N
 
 | | The crystal structure of Pyrococcus furiosus phosphoglucose isomerase with bound 5-phospho-D-arabinonate and Manganese | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, MANGANESE (II) ION | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-16 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
1X8E
 
 | | Crystal structure of Pyrococcus furiosus phosphoglucose isomerase free enzyme | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
2GC3
 
 | |
2GC1
 
 | |
2GC0
 
 | |
2GC2
 
 | |
3I9V
 
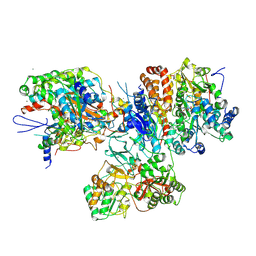 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, oxidized, 2 mol/ASU | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-13 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
3IAS
 
 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, oxidized, 4 mol/ASU, re-refined to 3.15 angstrom resolution | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-14 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
3IAM
 
 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, reduced, 2 mol/ASU, with bound NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-14 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
3ZT9
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, SERINE PHOSPHATASE | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
4HEA
 
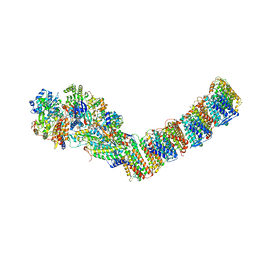 | | Crystal structure of the entire respiratory complex I from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Baradaran, R, Berrisford, J.M, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3027 Å) | | Cite: | Crystal structure of the entire respiratory complex I.
Nature, 494, 2013
|
|
4HE8
 
 | | Crystal structure of the membrane domain of respiratory complex I from Thermus thermophilus | | Descriptor: | NADH-quinone oxidoreductase subunit 10, NADH-quinone oxidoreductase subunit 11, NADH-quinone oxidoreductase subunit 12, ... | | Authors: | Baradaran, R, Berrisford, J.M, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the entire respiratory complex I.
Nature, 494, 2013
|
|
3ZTA
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZTB
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, IODIDE ION | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3ZXN
 
 | | Moorella thermoacetica RsbS S58E | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN, THIOCYANATE ION | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-08-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
3DD5
 
 | | Glomerella cingulata E600-cutinase complex | | Descriptor: | Cutinase, DIETHYL PHOSPHONATE | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-05 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DCN
 
 | | Glomerella cingulata apo cutinase | | Descriptor: | Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DEA
 
 | | Glomerella cingulata PETFP-cutinase complex | | Descriptor: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
