3CKY
 
 | |
2VUN
 
 | | The Crystal Structure of Enamidase at 1.9 A Resolution - A new Member of the Amidohydrolase Superfamily | | Descriptor: | CHLORIDE ION, ENAMIDASE, FE (III) ION, ... | | Authors: | Kress, D, Alhapel, A, Pierik, A.J, Essen, L.-O. | | Deposit date: | 2008-05-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Enamidase: A Bifunctional Enzyme of the Nicotinate Catabolism.
J.Mol.Biol., 384, 2008
|
|
2HES
 
 | | Cytosolic Iron-sulphur Assembly Protein- 1 | | Descriptor: | CALCIUM ION, Ydr267cp | | Authors: | Srinivasan, V, Michel, H, Lill, R, Daili, J.A.N, Pierik, A.J. | | Deposit date: | 2006-06-22 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Yeast WD40 Domain Protein Cia1, a Component Acting Late in Iron-Sulfur Protein Biogenesis.
Structure, 15, 2007
|
|
3HRD
 
 | | Crystal structure of nicotinate dehydrogenase | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wagener, N, Pierik, A.J, Hille, R, Dobbek, H. | | Deposit date: | 2009-06-09 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mo-Se active site of nicotinate dehydrogenase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ZXS
 
 | | Cryptochrome B from Rhodobacter sphaeroides | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, CRYPTOCHROME B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Geisselbrecht, Y, Fruhwirth, S, Pierik, A.J, Klug, G, Essen, L.-O. | | Deposit date: | 2011-08-15 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cryb from Rhodobacter Sphaeroides: A Unique Class of Cryptochromes with New Cofactors.
Embo Rep., 13, 2012
|
|
4D4O
 
 | | Crystal Structure of the Kti11 Kti13 heterodimer Spacegroup P64 | | Descriptor: | FE (III) ION, PROTEIN ATS1, DIPHTHAMIDE BIOSYNTHESIS PROTEIN 3, ... | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
4D4P
 
 | | Crystal Structure of the Kti11 Kti13 heterodimer Spacegroup P65 | | Descriptor: | FE (III) ION, PROTEIN ATS1, DIPHTHAMIDE BIOSYNTHESIS PROTEIN 3, ... | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
7NPA
 
 | |
7NP8
 
 | |
4OMF
 
 | | The F420-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member | | Descriptor: | CHLORIDE ION, F420-reducing hydrogenase, subunit alpha, ... | | Authors: | Vitt, S, Ma, K, Warkentin, E, Moll, J, Pierik, A, Shima, S, Ermler, U. | | Deposit date: | 2014-01-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The F420-Reducing [NiFe]-Hydrogenase Complex from Methanothermobacter marburgensis, the First X-ray Structure of a Group 3 Family Member.
J.Mol.Biol., 426, 2014
|
|
3G7K
 
 | |
4D4Q
 
 | | Crystal Structure of Kti13/AtS1 | | Descriptor: | PROTEIN ATS1 | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
4MYH
 
 | |
4MYC
 
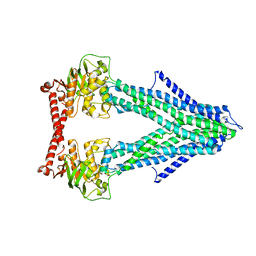 | | Structure of the mitochondrial ABC transporter, Atm1 | | Descriptor: | Iron-sulfur clusters transporter ATM1, mitochondrial | | Authors: | Srinivasan, V. | | Deposit date: | 2013-09-27 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1.
Science, 343, 2014
|
|
6QKR
 
 | |
6QKG
 
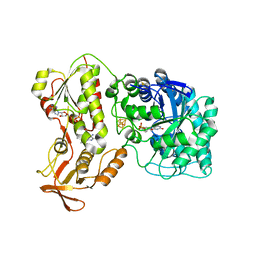 | | 2-Naphthoyl-CoA Reductase(NCR) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kayastha, K, Ermler, U. | | Deposit date: | 2019-01-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low potential enzymatic hydride transfer via highly cooperative and inversely functionalized flavin cofactors.
Nat Commun, 10, 2019
|
|
6QKX
 
 | |
1ERX
 
 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 COMPLEXED WITH NO | | Descriptor: | 5,8-DIMETHYL-1,2,3,4-TETRAVINYLPORPHINE-6,7-DIPROPIONIC ACID FERROUS COMPLEX, CITRIC ACID, NITRIC OXIDE, ... | | Authors: | Weichsel, A, Andersen, J.F, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-03 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nitric oxide binding to nitrophorin 4 induces complete distal pocket burial.
Nat.Struct.Biol., 7, 2000
|
|
