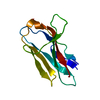
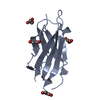
Entry Database : PDB / ID : 6o5kTitle Murine TRIM28 Bbox1 domain Transcription intermediary factor 1-beta Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Mus musculus (house mouse)Method / / / Resolution : 1.6 Å Authors Sun, Y. / Keown, J.R. / Goldstone, D.C. Funding support Organization Grant number Country Royal Society of New Zealand
Journal : J.Mol.Biol. / Year : 2019Title : A Dissection of Oligomerization by the TRIM28 Tripartite Motif and the Interaction with Members of the Krab-ZFP Family.Authors : Sun, Y. / Keown, J.R. / Black, M.M. / Raclot, C. / Demarais, N. / Trono, D. / Turelli, P. / Goldstone, D.C. History Deposition Mar 3, 2019 Deposition site / Processing site Revision 1.0 Jun 19, 2019 Provider / Type Revision 1.1 Jul 3, 2019 Group / Database references / Category Item / _citation.page_first / _citation.page_lastRevision 1.2 Mar 13, 2024 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / struct_ncs_dom_lim Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords TRANSCRIPTION / B-box KAP1 TIF1beta Krab-ZFP Co-repressor
TRANSCRIPTION / B-box KAP1 TIF1beta Krab-ZFP Co-repressor Function and homology information
Function and homology information chromo shadow domain binding / SUMO transferase activity / protein sumoylation /
chromo shadow domain binding / SUMO transferase activity / protein sumoylation /  epithelial to mesenchymal transition / positive regulation of DNA binding /
epithelial to mesenchymal transition / positive regulation of DNA binding /  heterochromatin /
heterochromatin /  embryo implantation / positive regulation of DNA repair / promoter-specific chromatin binding /
embryo implantation / positive regulation of DNA repair / promoter-specific chromatin binding /  euchromatin / RING-type E3 ubiquitin transferase / positive regulation of protein import into nucleus / RNA polymerase II transcription regulator complex / ubiquitin-protein transferase activity / transcription corepressor activity /
euchromatin / RING-type E3 ubiquitin transferase / positive regulation of protein import into nucleus / RNA polymerase II transcription regulator complex / ubiquitin-protein transferase activity / transcription corepressor activity /  ubiquitin protein ligase activity / chromatin organization / positive regulation of protein binding / in utero embryonic development / protein autophosphorylation /
ubiquitin protein ligase activity / chromatin organization / positive regulation of protein binding / in utero embryonic development / protein autophosphorylation /  transcription coactivator activity /
transcription coactivator activity /  protein kinase activity /
protein kinase activity /  protein phosphorylation /
protein phosphorylation /  DNA repair /
DNA repair /  innate immune response / negative regulation of DNA-templated transcription /
innate immune response / negative regulation of DNA-templated transcription /  chromatin binding /
chromatin binding /  ubiquitin protein ligase binding /
ubiquitin protein ligase binding /  chromatin / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / protein-containing complex /
chromatin / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / protein-containing complex /  DNA binding / zinc ion binding /
DNA binding / zinc ion binding /  nucleoplasm /
nucleoplasm /  nucleus
nucleus
 Mus musculus (house mouse)
Mus musculus (house mouse) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  SAD / Resolution: 1.6 Å
SAD / Resolution: 1.6 Å  Authors
Authors New Zealand, 1items
New Zealand, 1items  Citation
Citation Journal: J.Mol.Biol. / Year: 2019
Journal: J.Mol.Biol. / Year: 2019 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6o5k.cif.gz
6o5k.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6o5k.ent.gz
pdb6o5k.ent.gz PDB format
PDB format 6o5k.json.gz
6o5k.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/o5/6o5k
https://data.pdbj.org/pub/pdb/validation_reports/o5/6o5k ftp://data.pdbj.org/pub/pdb/validation_reports/o5/6o5k
ftp://data.pdbj.org/pub/pdb/validation_reports/o5/6o5k Links
Links Assembly
Assembly
 Components
Components
 Mus musculus (house mouse) / Gene: Trim28, Kap1, Krip1, Tif1b / Plasmid: pET-49+(b)-MBP / Production host:
Mus musculus (house mouse) / Gene: Trim28, Kap1, Krip1, Tif1b / Plasmid: pET-49+(b)-MBP / Production host: 
 Escherichia coli (E. coli) / Strain (production host): LOBSTR
Escherichia coli (E. coli) / Strain (production host): LOBSTR Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  Australian Synchrotron
Australian Synchrotron  / Beamline: MX1 / Wavelength: 0.9537 Å
/ Beamline: MX1 / Wavelength: 0.9537 Å : 0.9537 Å / Relative weight: 1
: 0.9537 Å / Relative weight: 1  Processing
Processing :
:  SAD / Resolution: 1.6→42.04 Å / Cor.coef. Fo:Fc: 0.966 / Cor.coef. Fo:Fc free: 0.957 / SU B: 5.574 / SU ML: 0.076 / Cross valid method: THROUGHOUT / ESU R: 0.083 / ESU R Free: 0.073 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
SAD / Resolution: 1.6→42.04 Å / Cor.coef. Fo:Fc: 0.966 / Cor.coef. Fo:Fc free: 0.957 / SU B: 5.574 / SU ML: 0.076 / Cross valid method: THROUGHOUT / ESU R: 0.083 / ESU R Free: 0.073 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller











 PDBj
PDBj



