| Entry | Database: PDB / ID: 5lne
|
|---|
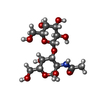
| Title | E. coli F9 pilus adhesin FmlH bound to the Thomsen-Friedenreich (TF) antigen |
|---|
 Components Components | Putative Fml fimbrial adhesin FmlD |
|---|
 Keywords Keywords | CELL ADHESION / fimbrial adhesin / lectin / O-glycan |
|---|
| Function / homology |  Function and homology information Function and homology information
Fimbrial-type adhesion domain / FimH, mannose-binding domain / FimH, mannose binding / : / Fimbrial-type adhesion domain superfamily / Adhesion domain superfamily / Immunoglobulin-like / Sandwich / Mainly BetaSimilarity search - Domain/homology Thomsen-Friedenreich antigen / NICKEL (II) ION / Hypothetical fimbrial-like protein ydeQ / Putative Fml fimbrial adhesin FmlDSimilarity search - Component |
|---|
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å MOLECULAR REPLACEMENT / Resolution: 2.2 Å |
|---|
 Authors Authors | Ruer, S. / Conover, M.S. / Kalas, V. / Taganna, J. / De Greve, H. / Pinkner, J.S. / Dodson, K.W. / Hultgren, S.J. / Remaut, H. |
|---|
| Funding support |  Belgium, Belgium,  United States, 2items United States, 2items | Organization | Grant number | Country |
|---|
| FWO | G030411N |  Belgium Belgium | | National Institutes of Health | DK051406 |  United States United States |
|
|---|
 Citation Citation |  Journal: Cell Host Microbe / Year: 2016 Journal: Cell Host Microbe / Year: 2016
Title: Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection.
Authors: Conover, M.S. / Ruer, S. / Taganna, J. / Kalas, V. / De Greve, H. / Pinkner, J.S. / Dodson, K.W. / Remaut, H. / Hultgren, S.J. |
|---|
| History | | Deposition | Aug 4, 2016 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Oct 5, 2016 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Oct 26, 2016 | Group: Database references |
|---|
| Revision 1.2 | Jan 31, 2018 | Group: Author supporting evidence / Category: pdbx_audit_support / Item: _pdbx_audit_support.funding_organization |
|---|
| Revision 2.0 | Jul 29, 2020 | Group: Atomic model / Data collection ...Atomic model / Data collection / Derived calculations / Structure summary
Category: atom_site / atom_site_anisotrop ...atom_site / atom_site_anisotrop / chem_comp / entity / entity_name_com / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_molecule_features / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / struct_asym / struct_conn / struct_site / struct_site_gen
Item: _atom_site.auth_asym_id / _atom_site.auth_atom_id ..._atom_site.auth_asym_id / _atom_site.auth_atom_id / _atom_site.auth_seq_id / _atom_site.label_asym_id / _atom_site.label_atom_id / _atom_site.label_entity_id / _atom_site_anisotrop.pdbx_auth_asym_id / _atom_site_anisotrop.pdbx_auth_atom_id / _atom_site_anisotrop.pdbx_auth_seq_id / _atom_site_anisotrop.pdbx_label_asym_id / _atom_site_anisotrop.pdbx_label_atom_id / _chem_comp.name / _chem_comp.type / _pdbx_struct_assembly_gen.asym_id_list / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id
Description: Carbohydrate remediation / Provider: repository / Type: Remediation |
|---|
| Revision 2.1 | Oct 16, 2024 | Group: Data collection / Database references ...Data collection / Database references / Derived calculations / Refinement description / Structure summary
Category: chem_comp / chem_comp_atom ...chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_entry_details / pdbx_modification_feature / struct_conn / struct_ncs_dom_lim
Item: _chem_comp.pdbx_synonyms / _database_2.pdbx_DOI ..._chem_comp.pdbx_synonyms / _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.pdbx_leaving_atom_flag / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.2 Å
MOLECULAR REPLACEMENT / Resolution: 2.2 Å  Authors
Authors Belgium,
Belgium,  United States, 2items
United States, 2items  Citation
Citation Journal: Cell Host Microbe / Year: 2016
Journal: Cell Host Microbe / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5lne.cif.gz
5lne.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5lne.ent.gz
pdb5lne.ent.gz PDB format
PDB format 5lne.json.gz
5lne.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ln/5lne
https://data.pdbj.org/pub/pdb/validation_reports/ln/5lne ftp://data.pdbj.org/pub/pdb/validation_reports/ln/5lne
ftp://data.pdbj.org/pub/pdb/validation_reports/ln/5lne Links
Links Assembly
Assembly

 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I03 / Wavelength: 0.97631 Å
/ Beamline: I03 / Wavelength: 0.97631 Å Processing
Processing MOLECULAR REPLACEMENT / Resolution: 2.2→59.4 Å / Cor.coef. Fo:Fc: 0.959 / Cor.coef. Fo:Fc free: 0.935 / SU B: 11.792 / SU ML: 0.144 / Cross valid method: THROUGHOUT / ESU R: 0.258 / ESU R Free: 0.198 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MOLECULAR REPLACEMENT / Resolution: 2.2→59.4 Å / Cor.coef. Fo:Fc: 0.959 / Cor.coef. Fo:Fc free: 0.935 / SU B: 11.792 / SU ML: 0.144 / Cross valid method: THROUGHOUT / ESU R: 0.258 / ESU R Free: 0.198 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller











 PDBj
PDBj