+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1feo | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of omega-conotoxin MVIIA with C-terminal Gly | ||||||
 Components Components | OMEGA-CONOTOXIN MVIIA-GLY | ||||||
 Keywords Keywords | TOXIN / beta sheet / disulfide knot | ||||||
| Function / homology |  Function and homology information Function and homology informationhost cell presynaptic membrane / ion channel inhibitor activity / calcium channel regulator activity / toxin activity / extracellular region Similarity search - Function | ||||||
| Method | SOLUTION NMR / simulated annealing in torsion angle space | ||||||
 Authors Authors | Goldenberg, D.P. / Koehn, R.E. / Gilbert, D.E. / Wagner, G. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2001 Journal: Protein Sci. / Year: 2001Title: Solution structure and backbone dynamics of an omega-conotoxin precursor Authors: Goldenberg, D.P. / Koehn, R.E. / Gilbert, D.E. / Wagner, G. #1:  Journal: Biochemistry / Year: 1996 Journal: Biochemistry / Year: 1996Title: Folding of omega-conotoxins. 2. Influence of precursor sequences and protein disulfide isomerase Authors: Price-Carter, M. / Gray, W.R. / Goldenberg, D.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1feo.cif.gz 1feo.cif.gz | 124.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1feo.ent.gz pdb1feo.ent.gz | 88.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1feo.json.gz 1feo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fe/1feo https://data.pdbj.org/pub/pdb/validation_reports/fe/1feo ftp://data.pdbj.org/pub/pdb/validation_reports/fe/1feo ftp://data.pdbj.org/pub/pdb/validation_reports/fe/1feo | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 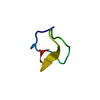
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 2709.268 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: Chemically synthesized sequence based on c-DNA from Conus magus. 15N-labeled sample was produced in Escherichia coli using a synthetic gene. References: UniProt: P05484 |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | pH: 6 / Pressure: ambient / Temperature: 283 K | |||||||||
| Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: simulated annealing in torsion angle space / Software ordinal: 1 Details: The structures are based on 249 non-reduncant NOE-derived distance restraints, 9 distance restraints from the three disulfide bonds, 16 distance restraints from hydrogen bonds, 19 dihedral angle restraints. | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 50 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller













 PDBj
PDBj NMRPipe
NMRPipe