[English] 日本語
 Yorodumi
Yorodumi- EMDB-1634: Structural analysis of substrate binding by the TatBC component o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1634 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. | |||||||||
 Map data Map data | This is a 3D map of a TatBC complex with one sufI substrate bound. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | twin arginine / Tat / protein transport / blue native PAGE / single particle electron microscopy | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining | |||||||||
 Authors Authors | Tarry MJ / Schaefer E / Chen S / Buchanan G / Greene NP / Lea SM / Palmer T / Saibil HR / Berks BC | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2009 Journal: Proc Natl Acad Sci U S A / Year: 2009Title: Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Authors: Michael J Tarry / Eva Schäfer / Shuyun Chen / Grant Buchanan / Nicholas P Greene / Susan M Lea / Tracy Palmer / Helen R Saibil / Ben C Berks /  Abstract: The Tat system transports folded proteins across the bacterial cytoplasmic membrane and the thylakoid membrane of plant chloroplasts. In Escherichia coli substrate proteins initially bind to the ...The Tat system transports folded proteins across the bacterial cytoplasmic membrane and the thylakoid membrane of plant chloroplasts. In Escherichia coli substrate proteins initially bind to the integral membrane TatBC complex which then recruits the protein TatA to effect translocation. Overproduction of TatBC and the substrate protein SufI in the absence of TatA led to the accumulation of TatBC-SufI complexes that could be purified using an affinity tag on the substrate. Three-dimensional structures of the TatBC-SufI complexes and unliganded TatBC were obtained by single-particle electron microscopy and random conical tilt reconstruction. Comparison of the structures shows that substrate molecules bind on the periphery of the TatBC complex and that substrate binding causes a significant reduction in diameter of the TatBC part of the complex. Although the TatBC complex contains multiple copies of the signal peptide-binding TatC protomer, purified TatBC-SufI complexes contain only 1 or 2 SufI molecules. Where 2 substrates are present in the TatBC-SufI complex, they are bound at adjacent sites. These observations imply that only certain TatC protomers within the complex interact with substrate or that there is a negative cooperativity of substrate binding. Similar TatBC-substrate complexes can be generated by an alternative in vitro reconstitution method and using a different substrate protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1634.map.gz emd_1634.map.gz | 40.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1634-v30.xml emd-1634-v30.xml emd-1634.xml emd-1634.xml | 8.4 KB 8.4 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1634.gif EMD-1634.gif | 26.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1634 http://ftp.pdbj.org/pub/emdb/structures/EMD-1634 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1634 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1634 | HTTPS FTP |
-Validation report
| Summary document |  emd_1634_validation.pdf.gz emd_1634_validation.pdf.gz | 187.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1634_full_validation.pdf.gz emd_1634_full_validation.pdf.gz | 186.9 KB | Display | |
| Data in XML |  emd_1634_validation.xml.gz emd_1634_validation.xml.gz | 4.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1634 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1634 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1634 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1634 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1634.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1634.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a 3D map of a TatBC complex with one sufI substrate bound. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
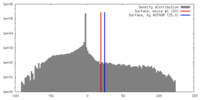
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : tatBC structure with one sufI substrate bound
| Entire | Name: tatBC structure with one sufI substrate bound |
|---|---|
| Components |
|
-Supramolecule #1000: tatBC structure with one sufI substrate bound
| Supramolecule | Name: tatBC structure with one sufI substrate bound / type: sample / ID: 1000 / Number unique components: 3 |
|---|
-Macromolecule #1: twin arginine transporter
| Macromolecule | Name: twin arginine transporter / type: protein_or_peptide / ID: 1 / Name.synonym: tat / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 0.014 |
|---|---|
| Staining | Type: NEGATIVE Details: negatively stained with 2% (wt/vol) uranyl acetate on glow discharged, continuous carbon-coated 300 mesh copper grids (Agar Scientific). |
| Grid | Details: 300 |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm Details: Micrographs were digitized on a Zeiss SCAI scanner at a pixel size of 7 micron, corresponding to 1.667 angstroms on the specimen. Subsequently, adjacent pixels were 3 x 3 averaged to yield a ...Details: Micrographs were digitized on a Zeiss SCAI scanner at a pixel size of 7 micron, corresponding to 1.667 angstroms on the specimen. Subsequently, adjacent pixels were 3 x 3 averaged to yield a pixel size of 5 angstroms. |
| Tilt angle min | 0 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: OTHER / Tilt angle max: 50 |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Software - Name: SPIDER Details: The tilted images were corrected for the effects of the contrast transfer function (CTF) by phase flipping, taking into account the defocus gradient across the micrographs and the position ...Details: The tilted images were corrected for the effects of the contrast transfer function (CTF) by phase flipping, taking into account the defocus gradient across the micrographs and the position of each particle. Images were processed using SPIDER version 11.12 and 15.06. Three-dimensional reconstruction was performed by the random conical tilt method in SPIDER. The particles were windowed into 64 x 64 pixel boxes. Number images used: 1556 |
|---|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera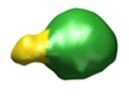









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)