[English] 日本語
 Yorodumi
Yorodumi- EMDB-1049: A cellular receptor of human rhinovirus type 2, the very-low-dens... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1049 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A cellular receptor of human rhinovirus type 2, the very-low-density lipoprotein receptor, binds to two neighboring proteins of the viral capsid. | |||||||||
 Map data Map data | This is a 3D reconstruction of HRV2/MBP-VLDLR(-123) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Human rhinovirus 2 Human rhinovirus 2 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Neumann E / Moser R / Snyers L / Blaas D / Hewat EA | |||||||||
 Citation Citation |  Journal: J Virol / Year: 2003 Journal: J Virol / Year: 2003Title: A cellular receptor of human rhinovirus type 2, the very-low-density lipoprotein receptor, binds to two neighboring proteins of the viral capsid. Authors: Emmanuelle Neumann / Rosita Moser / Luc Snyers / Dieter Blaas / Elizabeth A Hewat /  Abstract: The very-low-density lipoprotein receptor (VLDL-R) is a receptor for the minor-group human rhinoviruses (HRVs). Only two of the eight binding repeats of the VLDL-R bind to HRV2, and their footprints ...The very-low-density lipoprotein receptor (VLDL-R) is a receptor for the minor-group human rhinoviruses (HRVs). Only two of the eight binding repeats of the VLDL-R bind to HRV2, and their footprints describe an annulus on the dome at each fivefold axis. By studying the complex formed between a selection of soluble fragments of the VLDL-R and HRV2, we demonstrate that it is the second and third repeats that bind. We also show that artificial concatemers of the same repeat can bind to HRV2 with the same footprint as that for the native receptor. In a 16-A-resolution cryoelectron microscopy map of HRV2 in complex with the VLDL-R, the individual repeats are defined. The third repeat is strongly bound to charged and polar residues of the HI and BC loops of viral protein 1 (VP1), while the second repeat is more weakly bound to the neighboring VP1. The footprint of the strongly bound third repeat extends down the north side of the canyon. Since the receptor molecule can bind to two adjacent copies of VP1, we suggest that the bound receptor "staples" the VP1s together and must be detached before release of the RNA can occur. When the receptor is bound to neighboring sites on HRV2, steric hindrance prevents binding of the second repeat. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1049.map.gz emd_1049.map.gz | 15.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1049-v30.xml emd-1049-v30.xml emd-1049.xml emd-1049.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1049.gif 1049.gif | 40 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1049 http://ftp.pdbj.org/pub/emdb/structures/EMD-1049 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1049 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1049 | HTTPS FTP |
-Validation report
| Summary document |  emd_1049_validation.pdf.gz emd_1049_validation.pdf.gz | 247.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_1049_full_validation.pdf.gz emd_1049_full_validation.pdf.gz | 246.6 KB | Display | |
| Data in XML |  emd_1049_validation.xml.gz emd_1049_validation.xml.gz | 5.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1049 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1049 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1049 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1049 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1049.map.gz / Format: CCP4 / Size: 30.9 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_1049.map.gz / Format: CCP4 / Size: 30.9 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a 3D reconstruction of HRV2/MBP-VLDLR(-123) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.76 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
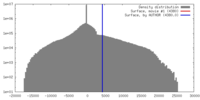
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human Rhinovirus 2 in complex with its cellular receptor, VLDL-R
| Entire | Name: Human Rhinovirus 2 in complex with its cellular receptor, VLDL-R |
|---|---|
| Components |
|
-Supramolecule #1000: Human Rhinovirus 2 in complex with its cellular receptor, VLDL-R
| Supramolecule | Name: Human Rhinovirus 2 in complex with its cellular receptor, VLDL-R type: sample / ID: 1000 / Number unique components: 2 |
|---|
-Supramolecule #1: Human rhinovirus 2
| Supramolecule | Name: Human rhinovirus 2 / type: virus / ID: 1 / NCBI-ID: 12130 / Sci species name: Human rhinovirus 2 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Virus shell | Shell ID: 1 / Diameter: 300 Å |
-Supramolecule #2: Cellular Receptor
| Supramolecule | Name: Cellular Receptor / type: organelle_or_cellular_component / ID: 2 / Name.synonym: Very Low Density Lipoprotein - Receptor Details: The VLDL-R consists of eight imperfect ligand-binding repeats of approximately 40 amino acids at its N-terminus. Recombinant VLDL-minireceptors encompassing different ligand-binding repeats ...Details: The VLDL-R consists of eight imperfect ligand-binding repeats of approximately 40 amino acids at its N-terminus. Recombinant VLDL-minireceptors encompassing different ligand-binding repeats are here expressed with the first three repeats + a Maltose Binding Protein fused to the N-terminus + a Hexa-his-tag fused to the C-terminus. Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Tissue: muscle Homo sapiens (human) / synonym: Human / Tissue: muscle |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.82 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 50 mM Tris-HCl for the virus 2 mM CaCl2, 20 mM Tris-HCL for the receptor fragment |
| Grid | Details: 300 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Method: Blot with filter paper for 1-2 seconds |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2010F |
|---|---|
| Alignment procedure | Legacy - Astigmatism: objective lens astigmatism was corrected at 300,000 times magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 7 µm / Number real images: 33 / Average electron dose: 15 e/Å2 / Details: Zeiss PhotoScan TD scanner |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 39700 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 1.4 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.18 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: Oxford cryo-holder CT3200 / Specimen holder model: OTHER |
- Image processing
Image processing
| Details | HRV2 and receptor fragment were incubated for 1 hour at 4 degree Celcius. |
|---|---|
| CTF correction | Details: CTFMIX |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.5 CUT-OFF Details: Methods for reconstructing density maps of "single" particles from cryoelectron micrographs to subnanometer resolution (Conway et al.,J Struct Biol. 1999 Dec 1;128(1):106-18.) Number images used: 912 |
-Atomic model buiding 1
| Software | Name: O |
|---|---|
| Details | Protocol: Manual docking. Fitting the X-RAY structures of HRV2 and the VLDL-R repeats to the cryo-electron microscope reconstructed density. Determination of the residues included in the footprints. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)