+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDF94 |
|---|---|
 Sample Sample | Insulin glulisine (Apidra), oligomeric composition
|
 Citation Citation |  Date: 2019 Jul Date: 2019 JulTitle: The quaternary structure of insulin glargine and glulisine under formulation conditions Authors: Nagel N / Graewert M / Gao M / Heyse W / Jeffries C / Svergun D |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #2919 |  Type: atomic / Comment: monomeric unit / Chi-square value: 1.03 / P-value: 0.574548  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|---|
| Model #2920 |  Type: atomic / Comment: dimeric unit / Chi-square value: 1.03 / P-value: 0.574548  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #2921 |  Type: atomic / Comment: hexameric unit / Chi-square value: 1.03 / P-value: 0.574548  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
| Model #2922 |  Type: atomic / Chi-square value: 1.03 / P-value: 0.574548  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
- Sample
Sample
 Sample Sample | Name: Insulin glulisine (Apidra), oligomeric composition / Specimen concentration: 3.49 mg/ml |
|---|---|
| Buffer | Name: Apidra formulation (per ml: 5 mg Sodium chloride, 3.15 mg m-Cresol, 6 mg Trometamol, 0.01 mg Polysorbate 20) pH: 7.3 |
| Entity #1567 | Type: protein / Description: Insulin glulisine / Formula weight: 5.811 / Num. of mol.: 6 Sequence: GIVEQCCTSI CSLYQLENYC NFVKQHLCGS HLVEALYLVC GERGFFYTPE T |
-Experimental information
| Beam | Instrument name: PETRA III EMBL P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.1241 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.1241 Å / Dist. spec. to detc.: 3.1 mm | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | |||||||||||||||||||||||||||||
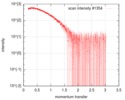
| Scan |
| |||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||
| Result | Comments: Here, Apidra measured at 3.49 mg/ml is displayed (formulation concentration). Measurements from 2 dilutions in placebo have been deposited in addition. Mixture analysis of the scattering ...Comments: Here, Apidra measured at 3.49 mg/ml is displayed (formulation concentration). Measurements from 2 dilutions in placebo have been deposited in addition. Mixture analysis of the scattering profiles was performed with the program Oligomer. The SAXS data show that Apidra® primarily consists of hexamers, which dissociate into monomers upon dilution. The presence of dimers was not required to fit the experimental data, and their volume fraction was always essentially zero. A small volume fraction is, however, also made up by larger dodecameric species. The distribution between monomer, dimer, hexamer and dodecamer is documented in the logfile. Calculation of the form factors is based on an internal model (Apidra.pdb) for hexamers and 3W80.pdb for dodecamers.
|
 Movie
Movie Controller
Controller


 SASDF94
SASDF94