[English] 日本語
 Yorodumi
Yorodumi- SASDB79: Basic domain of human telomeric repeat-binding factor 2 (TRF2) in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDB79 |
|---|---|
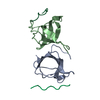
 Sample Sample | Basic domain of human telomeric repeat-binding factor 2 (TRF2) in complex with telomeric DNA duplex
|
| Function / homology |  Function and homology information Function and homology informationaxonal transport of messenger ribonucleoprotein complex / negative regulation of telomere single strand break repair / negative regulation of telomere maintenance via recombination / telomeric loop formation / negative regulation of telomere maintenance via semi-conservative replication / negative regulation of telomeric D-loop disassembly / negative regulation of telomere capping / protection from non-homologous end joining at telomere / RNA-templated DNA biosynthetic process / negative regulation of telomere maintenance ...axonal transport of messenger ribonucleoprotein complex / negative regulation of telomere single strand break repair / negative regulation of telomere maintenance via recombination / telomeric loop formation / negative regulation of telomere maintenance via semi-conservative replication / negative regulation of telomeric D-loop disassembly / negative regulation of telomere capping / protection from non-homologous end joining at telomere / RNA-templated DNA biosynthetic process / negative regulation of telomere maintenance / negative regulation of t-circle formation / telomeric D-loop disassembly / shelterin complex / Telomere C-strand synthesis initiation / regulation of telomere maintenance via telomerase / double-stranded telomeric DNA binding / Telomere C-strand (Lagging Strand) Synthesis / nuclear telomere cap complex / G-rich strand telomeric DNA binding / telomere capping / Processive synthesis on the C-strand of the telomere / Polymerase switching on the C-strand of the telomere / Removal of the Flap Intermediate from the C-strand / regulation of telomere maintenance / negative regulation of telomere maintenance via telomere lengthening / protein localization to chromosome, telomeric region / telomeric DNA binding / negative regulation of telomere maintenance via telomerase / positive regulation of telomere maintenance / negative regulation of cellular senescence / Telomere Extension By Telomerase / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / Recognition and association of DNA glycosylase with site containing an affected pyrimidine / Cleavage of the damaged pyrimidine / telomere maintenance / Inhibition of DNA recombination at telomere / Meiotic synapsis / male germ cell nucleus / DNA Damage/Telomere Stress Induced Senescence / cellular senescence / in utero embryonic development / chromosome, telomeric region / nuclear body / axon / protein-containing complex binding / enzyme binding / protein homodimerization activity / nucleoplasm / nucleus Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2017 Sep 13 Date: 2017 Sep 13Title: Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it Authors: Nečasová I / Janoušková E / Klumpler T |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDB79 SASDB79 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #1155 |  Type: dummy / Radius of dummy atoms: 2.25 A / Chi-square value: 1.061 / P-value: 0.955000  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: Basic domain of human telomeric repeat-binding factor 2 (TRF2) in complex with telomeric DNA duplex Specimen concentration: 2.9 mg/ml / Entity id: 569 / 570 |
|---|---|
| Buffer | Name: 20 mM Tris-HCl, 50 mM LiCl / pH: 7.5 |
| Entity #569 | Name: TRF2 / Type: protein Description: Basic domain of telomeric repeat-binding factor 2 Formula weight: 4.587 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q15554 Sequence: GPPGSMAGGG GSSDGSGRAA GRRASRSSGR ARRGRHEPGL GGPAERGAG |
| Entity #570 | Type: DNA / Description: telomere DNA duplex / Formula weight: 10.522 / Num. of mol.: 1 Sequence: GTTAGGGTTA GGGTTAGCAA TCCCAATCCC AATC |
-Experimental information
| Beam | Instrument name: CEITEC Rigaku BioSAXS-1000 / City: Brno / 国: Czech Republic  / Type of source: X-ray in house / Wavelength: 0.154 Å / Dist. spec. to detc.: 0.4791 mm / Type of source: X-ray in house / Wavelength: 0.154 Å / Dist. spec. to detc.: 0.4791 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 100K / Pixsize x: 172 mm | |||||||||||||||||||||||||||||||||
| Scan |
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result | Comments: The two-phase bead model of the protein-DNA complex was generated using MONSA refinement from SAXS data measured from the complex (top data-model fit) in parallel with data measured from ...Comments: The two-phase bead model of the protein-DNA complex was generated using MONSA refinement from SAXS data measured from the complex (top data-model fit) in parallel with data measured from the isolated telomeric DNA duplex (bottom data-model fit; refer to SASBDB entry SASDB89).
|
 Movie
Movie Controller
Controller