+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9izf | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of LPA1-Gi complex with LPA | |||||||||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / GPCR / SIGNALING PROTEIN | |||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to 1-oleoyl-sn-glycerol 3-phosphate / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Prostacyclin signalling through prostacyclin receptor / lysophosphatidic acid receptor activity / G alpha (z) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / positive regulation of smooth muscle cell chemotaxis ...cellular response to 1-oleoyl-sn-glycerol 3-phosphate / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Prostacyclin signalling through prostacyclin receptor / lysophosphatidic acid receptor activity / G alpha (z) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / positive regulation of smooth muscle cell chemotaxis / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / G beta:gamma signalling through BTK / Thromboxane signalling through TP receptor / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (s) signalling events / Lysosphingolipid and LPA receptors / lysophosphatidic acid binding / G-protein activation / Ca2+ pathway / G alpha (12/13) signalling events / Extra-nuclear estrogen signaling / G alpha (q) signalling events / Vasopressin regulates renal water homeostasis via Aquaporins / GPER1 signaling / negative regulation of cilium assembly / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / corpus callosum development / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / ADP signalling through P2Y purinoceptor 1 / G alpha (i) signalling events / bleb assembly / optic nerve development / cellular response to oxygen levels / oligodendrocyte development / regulation of synaptic vesicle cycle / negative regulation of cAMP/PKA signal transduction / regulation of metabolic process / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of Rho protein signal transduction / positive regulation of dendritic spine development / G-protein alpha-subunit binding / adenylate cyclase inhibitor activity / positive regulation of stress fiber assembly / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / neurogenesis / myelination / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / cerebellum development / regulation of mitotic spindle organization / dendritic shaft / cell chemotaxis / PDZ domain binding / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / electron transport chain / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 Similarity search - Function | |||||||||||||||||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)synthetic construct (others)  | |||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.14 Å | |||||||||||||||||||||||||||||||||||||||
 Authors Authors | Suzuki, S. / Nishikawa, K. / Kamegawa, A. / Hiroaki, Y. / Suzuki, H. / Fujiyoshi, Y. | |||||||||||||||||||||||||||||||||||||||
| Funding support |  Japan, 1items Japan, 1items
| |||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2025 Journal: J Struct Biol / Year: 2025Title: Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins. Authors: Shota Suzuki / Kotaro Tanaka / Akiko Kamegawa / Kouki Nishikawa / Hiroshi Suzuki / Atsunori Oshima / Yoshinori Fujiyoshi /  Abstract: Lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) are bioactive lysophospholipids derived from cell membranes that activate the endothelial differentiation gene family of G protein- ...Lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) are bioactive lysophospholipids derived from cell membranes that activate the endothelial differentiation gene family of G protein-coupled receptors. Activation of these receptors triggers multiple downstream signaling cascades through G proteins such as Gi/o, Gq/11, and G12/13. Therefore, LPA and S1P mediate several physiological processes, including cytoskeletal dynamics, neurite retraction, cell migration, cell proliferation, and intracellular ion fluxes. The basis for the G-protein coupling selectivity of EDG receptors, however, remains unknown. Here, we present cryo-electron microscopy structures of LPA-activated LPA1 in complexes with G, G, and G heterotrimers Comparison of the three LPA1-G protein structures shows clearly different conformations of intracellular loop 2 (ICL2) and ICL3 that are likely induced by the different Gα protein interfaces. Interestingly, this G-protein interface interaction is a common feature of LPA and S1P receptors. Our findings provide clues to understanding the promiscuity of G-protein coupling in the endothelial differentiation gene family. | |||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9izf.cif.gz 9izf.cif.gz | 276 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9izf.ent.gz pdb9izf.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  9izf.json.gz 9izf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/iz/9izf https://data.pdbj.org/pub/pdb/validation_reports/iz/9izf ftp://data.pdbj.org/pub/pdb/validation_reports/iz/9izf ftp://data.pdbj.org/pub/pdb/validation_reports/iz/9izf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  61031MC  9izgC  9izhC  61034  61035  61036 C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules ABC
| #2: Protein | Mass: 40446.047 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAI1 / Production host: Homo sapiens (human) / Gene: GNAI1 / Production host:  |
|---|---|
| #3: Protein | Mass: 41772.562 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: N-terminal His tag C-terminal GS linker and HiBiT tag Source: (gene. exp.)  Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:  |
| #4: Protein | Mass: 7729.947 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
-Protein / Antibody / Non-polymers , 3 types, 3 molecules RS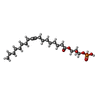

| #1: Protein | Mass: 73188.500 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: N-terminal HA signal sequence, FLAG tag C-terminal LgBiT Source: (gene. exp.)   Homo sapiens (human), (gene. exp.) synthetic construct (others) Homo sapiens (human), (gene. exp.) synthetic construct (others)Gene: cybC, LPAR1, EDG2, LPA1 / Production host:  |
|---|---|
| #5: Antibody | Mass: 26466.486 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.) synthetic construct (others) / Production host:  |
| #6: Chemical | ChemComp-NKP / ( |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Multiprotein complex / Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.4 |
| Specimen | Conc.: 15 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES / Details: This sample was monodisperse |
| Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 298 K |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: JEOL CRYO ARM 300 |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 71 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.14 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 178606 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj




































