+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of LPA1-G13 complex with LPA | |||||||||
 Map data Map data | main | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / SIGNALING PROTEIN / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to 1-oleoyl-sn-glycerol 3-phosphate / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Prostacyclin signalling through prostacyclin receptor / lysophosphatidic acid receptor activity / G alpha (z) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / positive regulation of smooth muscle cell chemotaxis ...cellular response to 1-oleoyl-sn-glycerol 3-phosphate / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Prostacyclin signalling through prostacyclin receptor / lysophosphatidic acid receptor activity / G alpha (z) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / positive regulation of smooth muscle cell chemotaxis / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / G beta:gamma signalling through BTK / Thromboxane signalling through TP receptor / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Lysosphingolipid and LPA receptors / G alpha (s) signalling events / lysophosphatidic acid binding / G-protein activation / Ca2+ pathway / G alpha (12/13) signalling events / Extra-nuclear estrogen signaling / G alpha (q) signalling events / Vasopressin regulates renal water homeostasis via Aquaporins / GPER1 signaling / negative regulation of cilium assembly / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / corpus callosum development / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / ADP signalling through P2Y purinoceptor 1 / G alpha (i) signalling events / bleb assembly / optic nerve development / cellular response to oxygen levels / oligodendrocyte development / regulation of synaptic vesicle cycle / negative regulation of cAMP/PKA signal transduction / regulation of metabolic process / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of Rho protein signal transduction / positive regulation of dendritic spine development / G-protein alpha-subunit binding / positive regulation of stress fiber assembly / neurogenesis / myelination / cerebellum development / dendritic shaft / cell chemotaxis / PDZ domain binding / electron transport chain / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / regulation of cell shape / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / negative regulation of neuron projection development / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / presynaptic membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) / Homo sapiens (human) / synthetic construct (others) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Suzuki S / Nishikawa K / Kamegawa A / Hiroaki Y / Suzuki H / Fujiyoshi Y | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2025 Journal: J Struct Biol / Year: 2025Title: Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins. Authors: Shota Suzuki / Kotaro Tanaka / Akiko Kamegawa / Kouki Nishikawa / Hiroshi Suzuki / Atsunori Oshima / Yoshinori Fujiyoshi /  Abstract: Lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) are bioactive lysophospholipids derived from cell membranes that activate the endothelial differentiation gene family of G protein- ...Lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) are bioactive lysophospholipids derived from cell membranes that activate the endothelial differentiation gene family of G protein-coupled receptors. Activation of these receptors triggers multiple downstream signaling cascades through G proteins such as Gi/o, Gq/11, and G12/13. Therefore, LPA and S1P mediate several physiological processes, including cytoskeletal dynamics, neurite retraction, cell migration, cell proliferation, and intracellular ion fluxes. The basis for the G-protein coupling selectivity of EDG receptors, however, remains unknown. Here, we present cryo-electron microscopy structures of LPA-activated LPA1 in complexes with G, G, and G heterotrimers Comparison of the three LPA1-G protein structures shows clearly different conformations of intracellular loop 2 (ICL2) and ICL3 that are likely induced by the different Gα protein interfaces. Interestingly, this G-protein interface interaction is a common feature of LPA and S1P receptors. Our findings provide clues to understanding the promiscuity of G-protein coupling in the endothelial differentiation gene family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_61033.map.gz emd_61033.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-61033-v30.xml emd-61033-v30.xml emd-61033.xml emd-61033.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_61033_fsc.xml emd_61033_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_61033.png emd_61033.png | 40.9 KB | ||
| Masks |  emd_61033_msk_1.map emd_61033_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-61033.cif.gz emd-61033.cif.gz | 7.4 KB | ||
| Others |  emd_61033_half_map_1.map.gz emd_61033_half_map_1.map.gz emd_61033_half_map_2.map.gz emd_61033_half_map_2.map.gz | 24.9 MB 24.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-61033 http://ftp.pdbj.org/pub/emdb/structures/EMD-61033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-61033 | HTTPS FTP |
-Related structure data
| Related structure data |  9izhMC  9izfC  9izgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_61033.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_61033.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.99 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_61033_msk_1.map emd_61033_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: A
| File | emd_61033_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: B
| File | emd_61033_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Multiprotein complex
| Entire | Name: Multiprotein complex |
|---|---|
| Components |
|
-Supramolecule #1: Multiprotein complex
| Supramolecule | Name: Multiprotein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Soluble cytochrome b562,Lysophosphatidic acid receptor 1,LgBiT tag
| Macromolecule | Name: Soluble cytochrome b562,Lysophosphatidic acid receptor 1,LgBiT tag type: protein_or_peptide / ID: 1 Details: N-terminal HA signal sequence, FLAG tag C-terminal LgBiT Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 73.1885 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDSP EMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LAAISTSIPV ISQPQFTAMN EPQCFYNESI A FFYNRSGK ...String: MKTIIALSYI FCLVFADYKD DDDKADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDSP EMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LAAISTSIPV ISQPQFTAMN EPQCFYNESI A FFYNRSGK HLATEWNTVS KLVMGLGITV CIFIMLANLL VMVAIYVNRR FHFPIYYLMA NLAAADFFAG LAYFYLMFNT GP NTRRLTV STWLLRQGLI DTSLTASVAN LLAIAIERHI TVFRMQLHTR MSNRRVVVVI VVIWTMAIVM GAIPSVGWNC ICD IENCSN MAPLYSDSYL VFWAIFNLVT FVVMVVLYAH IFGYVRQRTM RMSRHSSGPR RNRDTMMSLL KTVVIVLGAF IICW TPGLV LLLLDVCCPQ CDVLAYEKFF LLLAEFNSAM NPIIYSYRDK EMSATFRQIL CCQRSENPTG PTEGSDRSAS SLNHT ILAG VHSNDHSVVV FTLEDFVGDW EQTAAYNLDQ VLEQGGVSSL LQNLAVSVTP IQRIVRSGEN ALKIDIHVII PYEGLS ADQ MAQIEEVFKV VYPVDDHHFK VILPYGTLVI DGVTPNMLNY FGRPYEGIAV FDGKKITVTG TLWNGNKIID ERLITPD GS MLFRVTINS UniProtKB: Soluble cytochrome b562, Lysophosphatidic acid receptor 1 |
-Macromolecule #2: G protein subunit 13 (Gi2-mini-G13 chimera)
| Macromolecule | Name: G protein subunit 13 (Gi2-mini-G13 chimera) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.38615 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: STVSAEDKAA AERSKEIDKC LSREKTYVKR LVKILLLGAD NSGKSTFLKQ MRIIHGGSGG SGGTKGIHEY DFEIKNVPFK MVDVGGQRS ERKRWFECFD SVTSILFLVD SSDFNRLTES LNDFETIVNN RVFSNVSIIL FLNKTDLLEE KVQIVSIKDY F LEFEGDPH ...String: STVSAEDKAA AERSKEIDKC LSREKTYVKR LVKILLLGAD NSGKSTFLKQ MRIIHGGSGG SGGTKGIHEY DFEIKNVPFK MVDVGGQRS ERKRWFECFD SVTSILFLVD SSDFNRLTES LNDFETIVNN RVFSNVSIIL FLNKTDLLEE KVQIVSIKDY F LEFEGDPH CLRDVQKFLV ECFRNKRRDQ QQKPLYHHFT TAINTENARL IFRDVKDTIL HDNLKQLMLQ |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.772562 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHE NLYFQGSSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSA SQDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT G YLSCCRFL ...String: MHHHHHHHHE NLYFQGSSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSA SQDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT G YLSCCRFL DDNQIVTSSG DTTCALWDIE TGQQTTTFTG HTGDVMSLSL APDTRLFVSG ACDASAKLWD VREGMCRQTF TG HESDINA ICFFPNGNAF ATGSDDATCR LFDLRADQEL MTYSHDNIIC GITSVSFSKS GRLLLAGYDD FNCNVWDALK ADR AGVLAG HDNRVSCLGV TDDGMAVATG SWDSFLKIWN GGSGGGGSGG SSSGGVSGWR LFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.729947 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFCAIL UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 26.466486 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL K |
-Macromolecule #6: (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate
| Macromolecule | Name: (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate type: ligand / ID: 6 / Number of copies: 1 / Formula: NKP |
|---|---|
| Molecular weight | Theoretical: 436.52 Da |
| Chemical component information | 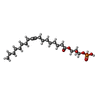 ChemComp-NKP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 15 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 71.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































