+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8yk0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human GPR156-Gi3 complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords | MEMBRANE PROTEIN / GPCR / class C / GPR156 / cryo-EM / homodimer / active state | ||||||
| Function / homology |  Function and homology information Function and homology informationstereocilium bundle organization / G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / negative regulation of adenylate cyclase activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma ...stereocilium bundle organization / G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / negative regulation of adenylate cyclase activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / GTP metabolic process / G alpha (i) signalling events / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / gamma-aminobutyric acid signaling pathway / G alpha (i) signalling events / spectrin binding / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / positive regulation of macroautophagy / photoreceptor outer segment / Adenylate cyclase inhibitory pathway / photoreceptor inner segment / cardiac muscle cell apoptotic process / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / centriolar satellite / ADP signalling through P2Y purinoceptor 12 / GDP binding / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / sensory perception of taste / signaling receptor complex adaptor activity / cell body / GTPase binding / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / midbody / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / cellular response to hypoxia / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / synapse / centrosome / dendrite / protein-containing complex binding / GTP binding / nucleolus / Golgi apparatus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)  | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.4 Å | ||||||
 Authors Authors | Ma, X.Y. / Chen, L.N. / Liao, M.H. / Zhang, L.Y. / Xi, K. / Guo, J.M. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular insights into the activation mechanism of GPR156 in maintaining auditory function. Authors: Xiangyu Ma / Li-Nan Chen / Menghui Liao / Liyan Zhang / Kun Xi / Jiamin Guo / Cangsong Shen / Dan-Dan Shen / Pengjun Cai / Qingya Shen / Jieyu Qi / Huibing Zhang / Shao-Kun Zang / Ying-Jun ...Authors: Xiangyu Ma / Li-Nan Chen / Menghui Liao / Liyan Zhang / Kun Xi / Jiamin Guo / Cangsong Shen / Dan-Dan Shen / Pengjun Cai / Qingya Shen / Jieyu Qi / Huibing Zhang / Shao-Kun Zang / Ying-Jun Dong / Luwei Miao / Jiao Qin / Su-Yu Ji / Yue Li / Jianfeng Liu / Chunyou Mao / Yan Zhang / Renjie Chai /  Abstract: The class C orphan G-protein-coupled receptor (GPCR) GPR156, which lacks the large extracellular region, plays a pivotal role in auditory function through G. Here, we firstly demonstrate that GPR156 ...The class C orphan G-protein-coupled receptor (GPCR) GPR156, which lacks the large extracellular region, plays a pivotal role in auditory function through G. Here, we firstly demonstrate that GPR156 with high constitutive activity is essential for maintaining auditory function, and further reveal the structural basis of the sustained role of GPR156. We present the cryo-EM structures of human apo GPR156 and the GPR156-G complex, unveiling a small extracellular region formed by extracellular loop 2 (ECL2) and the N-terminus. The GPR156 dimer in both apo state and G protein-coupled state adopt a transmembrane (TM)5/6-TM5/6 interface, indicating the high constitutive activity of GPR156 in the apo state. Furthermore, C-terminus in G-bound subunit of GPR156 plays a dual role in promoting G protein binding within G-bound subunit while preventing the G-free subunit from binding to additional G protein. Together, these results explain how GPR156 constitutive activity is maintained through dimerization and provide a mechanistic insight into the sustained role of GPR156 in maintaining auditory function. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8yk0.cif.gz 8yk0.cif.gz | 237 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8yk0.ent.gz pdb8yk0.ent.gz | 184.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8yk0.json.gz 8yk0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/yk/8yk0 https://data.pdbj.org/pub/pdb/validation_reports/yk/8yk0 ftp://data.pdbj.org/pub/pdb/validation_reports/yk/8yk0 ftp://data.pdbj.org/pub/pdb/validation_reports/yk/8yk0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  39356MC  8yjpC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Probable G-protein coupled receptor ... , 2 types, 2 molecules BA
| #1: Protein | Mass: 32797.066 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GPR156, GABABL, PGR28 / Production host: Homo sapiens (human) / Gene: GPR156, GABABL, PGR28 / Production host:  |
|---|---|
| #5: Protein | Mass: 35083.629 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GPR156, GABABL, PGR28 / Production host: Homo sapiens (human) / Gene: GPR156, GABABL, PGR28 / Production host:  |
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules DGC
| #2: Protein | Mass: 37198.656 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
|---|---|
| #3: Protein | Mass: 6289.240 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   |
| #4: Protein | Mass: 40078.512 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAI3 / Production host: Homo sapiens (human) / Gene: GNAI3 / Production host:  |
-Non-polymers , 2 types, 15 molecules 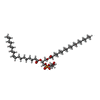


| #6: Chemical | ChemComp-MW9 / ( #7: Chemical | ChemComp-CLR / |
|---|
-Details
| Has ligand of interest | N |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: GPR156-Gi3 complex / Type: ORGANELLE OR CELLULAR COMPONENT / Entity ID: #1-#4 / Source: RECOMBINANT | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) |
| ||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1500 nm / Nominal defocus min: 700 nm / Cs: 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 52 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) |
- Processing
Processing
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3D reconstruction | Resolution: 2.4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 1153971 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




 PDBj
PDBj


















