+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human GPR156-Gi3 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / class C / GPR156 / cryo-EM / homodimer / active state / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationstereocilium bundle organization / G protein-coupled receptor heterodimeric complex / G protein-coupled GABA receptor activity / negative regulation of adenylate cyclase activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma ...stereocilium bundle organization / G protein-coupled receptor heterodimeric complex / G protein-coupled GABA receptor activity / negative regulation of adenylate cyclase activity / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / GTP metabolic process / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / gamma-aminobutyric acid signaling pathway / alkylglycerophosphoethanolamine phosphodiesterase activity / positive regulation of macroautophagy / photoreceptor outer segment / Adenylate cyclase inhibitory pathway / cardiac muscle cell apoptotic process / photoreceptor inner segment / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / ADP signalling through P2Y purinoceptor 12 / GDP binding / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / sensory perception of taste / signaling receptor complex adaptor activity / positive regulation of cytosolic calcium ion concentration / retina development in camera-type eye / cell body / GTPase binding / midbody / G alpha (i) signalling events / cellular response to hypoxia / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / synapse / dendrite / centrosome / GTP binding / protein-containing complex binding / nucleolus / Golgi apparatus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Ma XY / Chen LN / Liao MH / Zhang LY / Xi K / Guo JM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular insights into the activation mechanism of GPR156 in maintaining auditory function. Authors: Xiangyu Ma / Li-Nan Chen / Menghui Liao / Liyan Zhang / Kun Xi / Jiamin Guo / Cangsong Shen / Dan-Dan Shen / Pengjun Cai / Qingya Shen / Jieyu Qi / Huibing Zhang / Shao-Kun Zang / Ying-Jun ...Authors: Xiangyu Ma / Li-Nan Chen / Menghui Liao / Liyan Zhang / Kun Xi / Jiamin Guo / Cangsong Shen / Dan-Dan Shen / Pengjun Cai / Qingya Shen / Jieyu Qi / Huibing Zhang / Shao-Kun Zang / Ying-Jun Dong / Luwei Miao / Jiao Qin / Su-Yu Ji / Yue Li / Jianfeng Liu / Chunyou Mao / Yan Zhang / Renjie Chai /  Abstract: The class C orphan G-protein-coupled receptor (GPCR) GPR156, which lacks the large extracellular region, plays a pivotal role in auditory function through G. Here, we firstly demonstrate that GPR156 ...The class C orphan G-protein-coupled receptor (GPCR) GPR156, which lacks the large extracellular region, plays a pivotal role in auditory function through G. Here, we firstly demonstrate that GPR156 with high constitutive activity is essential for maintaining auditory function, and further reveal the structural basis of the sustained role of GPR156. We present the cryo-EM structures of human apo GPR156 and the GPR156-G complex, unveiling a small extracellular region formed by extracellular loop 2 (ECL2) and the N-terminus. The GPR156 dimer in both apo state and G protein-coupled state adopt a transmembrane (TM)5/6-TM5/6 interface, indicating the high constitutive activity of GPR156 in the apo state. Furthermore, C-terminus in G-bound subunit of GPR156 plays a dual role in promoting G protein binding within G-bound subunit while preventing the G-free subunit from binding to additional G protein. Together, these results explain how GPR156 constitutive activity is maintained through dimerization and provide a mechanistic insight into the sustained role of GPR156 in maintaining auditory function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39356.map.gz emd_39356.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39356-v30.xml emd-39356-v30.xml emd-39356.xml emd-39356.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39356.png emd_39356.png | 96.4 KB | ||
| Filedesc metadata |  emd-39356.cif.gz emd-39356.cif.gz | 7 KB | ||
| Others |  emd_39356_half_map_1.map.gz emd_39356_half_map_1.map.gz emd_39356_half_map_2.map.gz emd_39356_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39356 http://ftp.pdbj.org/pub/emdb/structures/EMD-39356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39356 | HTTPS FTP |
-Related structure data
| Related structure data |  8yk0MC  8yjpC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39356.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39356.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_39356_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_39356_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GPR156-Gi3 complex
| Entire | Name: GPR156-Gi3 complex |
|---|---|
| Components |
|
-Supramolecule #1: GPR156-Gi3 complex
| Supramolecule | Name: GPR156-Gi3 complex / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Probable G-protein coupled receptor 156
| Macromolecule | Name: Probable G-protein coupled receptor 156 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.797066 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RPLHDLCKTT ITSSHHSSKT ISSLSPVLLG IVWTFLSCGL LLILFFLAFT IHCRKNRIVK MSSPNLNIVT LLGSCLTYSS AYLFGIQDV LVGSSMETLI QTRLSMLCIG TSLVFGPILG KSWRLYKVFT QRVPDKRVII KDLQLLGLVA ALLMADVILL M TWVLTDPI ...String: RPLHDLCKTT ITSSHHSSKT ISSLSPVLLG IVWTFLSCGL LLILFFLAFT IHCRKNRIVK MSSPNLNIVT LLGSCLTYSS AYLFGIQDV LVGSSMETLI QTRLSMLCIG TSLVFGPILG KSWRLYKVFT QRVPDKRVII KDLQLLGLVA ALLMADVILL M TWVLTDPI QCLQILSVSM TVTGKDVSCT STSTHFCASR YSDVWIALIW GCKGLLLLYG AYLAGLTGHV SSPPVNQSLT IM VGVNLLV LAAGLLFVVT RYLHSWPNLV FGLTSGGIFV CTTTINCFIF IPQLKQWKAF E UniProtKB: Probable G-protein coupled receptor 156 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.198656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD ...String: ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLVS ASQDGKLIIW DSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG HTGYLSCCRF LDDNQIVTSS G DTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TFTGHESDIN AICFFPNGNA FA TGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKADRAGVLA GHDNRVSCLG VTD DGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 6.28924 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASIAQARKLV EQLKMEANID RIKVSKAAAD LMAYCEAHAK EDPLLTPVPA SENPFRE UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Guanine nucleotide-binding protein G(i) subunit alpha-3
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-3 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.078512 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SAEDKAAVER SKMIDRNLRE DGEKAAKEVK LLLLGAGESG KNTIVKQMKI IHEDGYSEDE CKQYKVVVYS NTIQSIIAII RAMGRLKID FGEAARADDA RQLFVLAGSA EEGVMTPELA GVIKRLWRDG GVQACFSRSR EYQLNDSASY YLNDLDRISQ S NYIPTQQD ...String: SAEDKAAVER SKMIDRNLRE DGEKAAKEVK LLLLGAGESG KNTIVKQMKI IHEDGYSEDE CKQYKVVVYS NTIQSIIAII RAMGRLKID FGEAARADDA RQLFVLAGSA EEGVMTPELA GVIKRLWRDG GVQACFSRSR EYQLNDSASY YLNDLDRISQ S NYIPTQQD VLRTRVKTTG IVETHFTFKD LYFKMFDVGA QRSERKKWIH CFEGVTAIIF CVALSDYDLV LAEDEEMNRM HA SMKLFDS ICNNKWFTET SIILFLNKKD LFEEKIKRSP LTICYPEYTG SNTYEEAAAY IQCQFEDLNR RKDTKEIYTH FTC STDTKN VQFVFDAVTD VIIKNNLKEC GLY UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-3 |
-Macromolecule #5: Probable G-protein coupled receptor 156
| Macromolecule | Name: Probable G-protein coupled receptor 156 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.083629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: RPLHDLCKTT ITSSHHSSKT ISSLSPVLLG IVWTFLSCGL LLILFFLAFT IHCRKNRIVK MSSPNLNIVT LLGSCLTYSS AYLFGIQDV LVGSSMETLI QTRLSMLCIG TSLVFGPILG KSWRLYKVFT QRVPDKRVII KDLQLLGLVA ALLMADVILL M TWVLTDPI ...String: RPLHDLCKTT ITSSHHSSKT ISSLSPVLLG IVWTFLSCGL LLILFFLAFT IHCRKNRIVK MSSPNLNIVT LLGSCLTYSS AYLFGIQDV LVGSSMETLI QTRLSMLCIG TSLVFGPILG KSWRLYKVFT QRVPDKRVII KDLQLLGLVA ALLMADVILL M TWVLTDPI QCLQILSVSM TVTGKDVSCT STSTHFCASR YSDVWIALIW GCKGLLLLYG AYLAGLTGHV SSPPVNQSLT IM VGVNLLV LAAGLLFVVT RYLHSWPNLV FGLTSGGIFV CTTTINCFIF IPQLKQWKAF EEENQTIRRM AKYFSTPNKS UniProtKB: Probable G-protein coupled receptor 156 |
-Macromolecule #6: (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24l...
| Macromolecule | Name: (21R,24R,27S)-24,27,28-trihydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaoctacosan-21-yl (9Z)-octadec-9-enoate type: ligand / ID: 6 / Number of copies: 4 / Formula: MW9 |
|---|---|
| Molecular weight | Theoretical: 777.06 Da |
| Chemical component information | 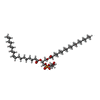 ChemComp-MW9: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 11 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































