[English] 日本語
 Yorodumi
Yorodumi- PDB-8q5i: Structure of Candida albicans 80S ribosome in complex with cephaeline -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8q5i | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of Candida albicans 80S ribosome in complex with cephaeline | ||||||
 Components Components |
| ||||||
 Keywords Keywords | RIBOSOME / Candida albicans / cephaeline | ||||||
| Function / homology |  Function and homology information Function and homology informationyeast-form cell wall / hyphal cell wall / preribosome / positive regulation of translational fidelity / pre-mRNA 5'-splice site binding / nonfunctional rRNA decay / preribosome, small subunit precursor / negative regulation of mRNA splicing, via spliceosome / preribosome, large subunit precursor / translational elongation ...yeast-form cell wall / hyphal cell wall / preribosome / positive regulation of translational fidelity / pre-mRNA 5'-splice site binding / nonfunctional rRNA decay / preribosome, small subunit precursor / negative regulation of mRNA splicing, via spliceosome / preribosome, large subunit precursor / translational elongation / 90S preribosome / ribosomal subunit export from nucleus / protein-RNA complex assembly / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal small subunit export from nucleus / DNA-(apurinic or apyrimidinic site) endonuclease activity / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / maintenance of translational fidelity / rRNA processing / large ribosomal subunit / ribosome biogenesis / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / nucleolus / cell surface / RNA binding / zinc ion binding / metal ion binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Candida albicans (yeast) Candida albicans (yeast)Synthetic construct (others) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.45 Å | ||||||
 Authors Authors | Kolosova, O. / Zgadzay, Y. / Stetsenko, A. / Atamas, A. / Guskov, A. / Yusupov, M. | ||||||
| Funding support |  France, 1items France, 1items
| ||||||
 Citation Citation |  Journal: Biopolym Cell / Year: 2023 Journal: Biopolym Cell / Year: 2023Title: Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy Authors: Kolosova, O. / Zgadzay, Y. / Stetsenko, A. / Atamas, A. / Wu, C. / Jenner, L. / Sachs, S.M. / Guskov, A. / Yusupov, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8q5i.cif.gz 8q5i.cif.gz | 4.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8q5i.ent.gz pdb8q5i.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8q5i.json.gz 8q5i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q5/8q5i https://data.pdbj.org/pub/pdb/validation_reports/q5/8q5i ftp://data.pdbj.org/pub/pdb/validation_reports/q5/8q5i ftp://data.pdbj.org/pub/pdb/validation_reports/q5/8q5i | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  18156MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-RNA chain , 6 types, 6 molecules 13410Ag
| #1: RNA chain | Mass: 1085306.250 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: GenBank: 1305241335 Candida albicans (yeast) / References: GenBank: 1305241335 |
|---|---|
| #2: RNA chain | Mass: 38943.129 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: GenBank: X16634.1 Candida albicans (yeast) / References: GenBank: X16634.1 |
| #3: RNA chain | Mass: 50683.906 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: GenBank: OQ195134.1 Candida albicans (yeast) / References: GenBank: OQ195134.1 |
| #4: RNA chain | Mass: 4461.723 Da / Num. of mol.: 1 / Source method: isolated from a natural source Details: The correct sequence should be GGCAGCAUAGCUCAGUUGGUUAGAGCGUAGGACUCAUAAGCCUAAGGUCACUGGUUCAAUUCCAGUUCUGCUACCA. Like GGCAG - gap - CUGCUACCA Source: (natural)  Candida albicans (yeast) Candida albicans (yeast) |
| #46: RNA chain | Mass: 575838.625 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: GenBank: XR_002086442.1 Candida albicans (yeast) / References: GenBank: XR_002086442.1 |
| #76: RNA chain | Mass: 1625.048 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) Synthetic construct (others) |
+60S ribosomal protein ... , 37 types, 37 molecules jklmnopqrstuvyz025678AAABACADAEAFAGAHAJ...
-Ribosomal protein ... , 10 types, 10 molecules wx9AIDEGKVY
| #18: Protein | Mass: 22685.850 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6BZ56 Candida albicans (yeast) / References: UniProt: A0A8H6BZ56 |
|---|---|
| #19: Protein | Mass: 21002.861 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6BUG0 Candida albicans (yeast) / References: UniProt: A0A8H6BUG0 |
| #28: Protein | Mass: 14215.550 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6F3X2 Candida albicans (yeast) / References: UniProt: A0A8H6F3X2 |
| #37: Protein | Mass: 14118.759 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6F6B7 Candida albicans (yeast) / References: UniProt: A0A8H6F6B7 |
| #49: Protein | Mass: 26917.314 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6BZ90 Candida albicans (yeast) / References: UniProt: A0A8H6BZ90 |
| #50: Protein | Mass: 27313.570 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6BYS1 Candida albicans (yeast) / References: UniProt: A0A8H6BYS1 |
| #52: Protein | Mass: 25319.922 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6BS29 Candida albicans (yeast) / References: UniProt: A0A8H6BS29 |
| #56: Protein | Mass: 21765.146 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6F1X2 Candida albicans (yeast) / References: UniProt: A0A8H6F1X2 |
| #66: Protein | Mass: 13366.540 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6BYJ9 Candida albicans (yeast) / References: UniProt: A0A8H6BYJ9 |
| #69: Protein | Mass: 16000.784 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Candida albicans (yeast) / References: UniProt: A0A8H6F0V0 Candida albicans (yeast) / References: UniProt: A0A8H6F0V0 |
+40S ribosomal protein ... , 23 types, 23 molecules BCFHIJLMOPQRSTUWXZbcdef
-Non-polymers , 6 types, 1545 molecules 
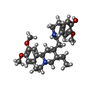









| #77: Chemical | ChemComp-SPK / | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #78: Chemical | | #79: Chemical | ChemComp-SPD / | #80: Chemical | ChemComp-MG / #81: Chemical | ChemComp-ZN / #82: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Candida albicans 80S ribosome in complex with cephaeline Type: RIBOSOME / Entity ID: #1-#75 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Candida albicans (yeast) Candida albicans (yeast) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2400 nm / Nominal defocus min: 900 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Version: 1.19rc4_4035: / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.45 Å / Resolution method: FSC 3 SIGMA CUT-OFF / Num. of particles: 254047 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj



































