[English] 日本語
 Yorodumi
Yorodumi- PDB-8owm: Crystal structure of glutamate dehydrogenase 2 from Arabidopsis t... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8owm | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of glutamate dehydrogenase 2 from Arabidopsis thaliana binding Ca, NAD and 2,2-dihydroxyglutarate | ||||||
 Components Components | Glutamate dehydrogenase 2 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / glutamic acid / calcium / NAD cofactor / nitrogen metabolism / 2 / 2-dihydroxyglutarate | ||||||
| Function / homology |  Function and homology information Function and homology informationL-glutamate dehydrogenase [NAD(P)+] activity / glutamate dehydrogenase [NAD(P)+] / L-glutamate dehydrogenase (NADP+) activity / L-glutamate dehydrogenase (NAD+) activity / plant-type vacuole / amino acid metabolic process / cobalt ion binding / copper ion binding / mitochondrion / zinc ion binding ...L-glutamate dehydrogenase [NAD(P)+] activity / glutamate dehydrogenase [NAD(P)+] / L-glutamate dehydrogenase (NADP+) activity / L-glutamate dehydrogenase (NAD+) activity / plant-type vacuole / amino acid metabolic process / cobalt ion binding / copper ion binding / mitochondrion / zinc ion binding / ATP binding / plasma membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.7 Å MOLECULAR REPLACEMENT / Resolution: 1.7 Å | ||||||
 Authors Authors | Grzechowiak, M. / Ruszkowski, M. | ||||||
| Funding support |  Poland, 1items Poland, 1items
| ||||||
 Citation Citation |  Journal: Plant Physiol Biochem / Year: 2023 Journal: Plant Physiol Biochem / Year: 2023Title: Structural and functional studies of Arabidopsis thaliana glutamate dehydrogenase isoform 2 demonstrate enzyme dynamics and identify its calcium binding site. Authors: Marta Grzechowiak / Joanna Sliwiak / Mariusz Jaskolski / Milosz Ruszkowski /  Abstract: Glutamate dehydrogenase (GDH) is an enzyme at the crossroad of plant nitrogen and carbon metabolism. GDH catalyzes the conversion of 2-oxoglutarate into glutamate (2OG → Glu), utilizing ammonia as ...Glutamate dehydrogenase (GDH) is an enzyme at the crossroad of plant nitrogen and carbon metabolism. GDH catalyzes the conversion of 2-oxoglutarate into glutamate (2OG → Glu), utilizing ammonia as cosubstrate and NADH as coenzyme. The GDH reaction is reversible, meaning that the NAD-dependent reaction (Glu → 2OG) releases ammonia. In Arabidopsis thaliana, three GDH isoforms exist, AtGDH1, AtGDH2, and AtGDH3. The subject of this work is AtGDH2. Previous reports have suggested that enzymes homologous to AtGDH2 contain a calcium-binding EF-hand motif located in the coenzyme binding domain. Here, we show that while AtGDH2 indeed does bind calcium, the binding occurs elsewhere and the region predicted to be the EF-hand motif has a completely different structure. As the true calcium binding site is > 20 Å away from the active site, it seems to play a structural, rather than catalytic role. We also performed comparative kinetic characterization of AtGDH1 and AtGDH2 using spectroscopic methods and isothermal titration calorimetry, to note that the isoenzymes generally exhibit similar behavior, with calcium having only a minor effect. However, the spatial and temporal changes in the gene expression profiles of the three AtGDH genes point to AtGDH2 as the most prevalent isoform. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8owm.cif.gz 8owm.cif.gz | 1007.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8owm.ent.gz pdb8owm.ent.gz | 829.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8owm.json.gz 8owm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ow/8owm https://data.pdbj.org/pub/pdb/validation_reports/ow/8owm ftp://data.pdbj.org/pub/pdb/validation_reports/ow/8owm ftp://data.pdbj.org/pub/pdb/validation_reports/ow/8owm | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8ownC  6yeiS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| Experimental dataset #1 | Data reference:  10.18150/II5MT4 / Data set type: diffraction image data / Metadata reference: 10.18150/II5MT4 10.18150/II5MT4 / Data set type: diffraction image data / Metadata reference: 10.18150/II5MT4 |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 6 molecules ABCDEF
| #1: Protein | Mass: 45027.188 Da / Num. of mol.: 6 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q38946, glutamate dehydrogenase [NAD(P)+] |
|---|
-Non-polymers , 8 types, 2156 molecules 


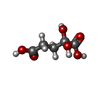











| #2: Chemical | ChemComp-NAD / #3: Chemical | ChemComp-GOL / #4: Chemical | ChemComp-EDO / #5: Chemical | ChemComp-U5C / #6: Chemical | ChemComp-CA / #7: Chemical | #8: Chemical | ChemComp-PEG / #9: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.87 Å3/Da / Density % sol: 57.16 % |
|---|---|
| Crystal grow | Temperature: 273 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: 0.12MMonosaccharides (0.02M D-Glucose; 0.02M D-Mannose; 0.02M D-Galactose; 0.02M L-Fucose; 0.02M D-Xylose; 0.02M N-Acetyl-D-Glucosamine ),0.1M Imidazole/MES buffer pH 6.5, 20% v/v Glycerol and 10% w/v PEG 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  PETRA III, EMBL c/o DESY PETRA III, EMBL c/o DESY  / Beamline: P13 (MX1) / Wavelength: 0.9762 Å / Beamline: P13 (MX1) / Wavelength: 0.9762 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Jun 21, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9762 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→65.7 Å / Num. obs: 318817 / % possible obs: 96.9 % / Redundancy: 3.55 % / Biso Wilson estimate: 23.46 Å2 / CC1/2: 0.998 / Rmerge(I) obs: 0.062 / Rrim(I) all: 0.075 / Net I/σ(I): 10.76 |
| Reflection shell | Resolution: 1.7→1.8 Å / Redundancy: 2.92 % / Rmerge(I) obs: 0.758 / Mean I/σ(I) obs: 1.32 / Num. unique obs: 50659 / CC1/2: 0.624 / Rrim(I) all: 0.927 / % possible all: 95.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6yei Resolution: 1.7→65.7 Å / SU ML: 0.1703 / Cross valid method: FREE R-VALUE / σ(F): 1.96 / Phase error: 18.7346 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.58 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.7→65.7 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj





