+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8g59 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | |||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / GPCR / GPR120 / complex / fatty acid hormones | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of somatostatin secretion / positive regulation of glucagon secretion / ghrelin secretion / Free fatty acid receptors / regulation of D-glucose transmembrane transport / taste receptor activity / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / phospholipase C-activating serotonin receptor signaling pathway ...negative regulation of somatostatin secretion / positive regulation of glucagon secretion / ghrelin secretion / Free fatty acid receptors / regulation of D-glucose transmembrane transport / taste receptor activity / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / phospholipase C-activating serotonin receptor signaling pathway / PLC beta mediated events / entrainment of circadian clock / regulation of platelet activation / hormone secretion / arrestin family protein binding / regulation of canonical Wnt signaling pathway / negative regulation of cytokine production / negative regulation of interleukin-1 beta production / glutamate receptor signaling pathway / ciliary membrane / white fat cell differentiation / phototransduction, visible light / postsynaptic cytosol / endocytic vesicle / photoreceptor outer segment / neuropeptide signaling pathway / positive regulation of osteoblast differentiation / brown fat cell differentiation / positive regulation of protein localization to cell cortex / Adenylate cyclase inhibitory pathway / T cell migration / D2 dopamine receptor binding / enzyme regulator activity / response to prostaglandin E / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / positive regulation of brown fat cell differentiation / cellular response to forskolin / regulation of mitotic spindle organization / GTPase activator activity / fatty acid binding / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / negative regulation of protein kinase activity / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / G protein-coupled receptor activity / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / response to peptide hormone / G-protein beta/gamma-subunit complex binding / centriolar satellite / negative regulation of inflammatory response / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / blood coagulation / GDP binding / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cold-induced thermogenesis / G protein activity / GTPase binding / positive regulation of cytosolic calcium ion concentration / Ca2+ pathway Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.64 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Mao, C. / Xiao, P. / Tao, X. / Qin, J. / He, Q. / Zhang, C. / Yu, X. / Zhang, Y. / Sun, J. | |||||||||||||||||||||||||||||||||
| Funding support | 1items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Unsaturated bond recognition leads to biased signal in a fatty acid receptor. Authors: Chunyou Mao / Peng Xiao / Xiao-Na Tao / Jiao Qin / Qing-Tao He / Chao Zhang / Sheng-Chao Guo / Ya-Qin Du / Li-Nan Chen / Dan-Dan Shen / Zhi-Shuai Yang / Han-Qiong Zhang / Shen-Ming Huang / ...Authors: Chunyou Mao / Peng Xiao / Xiao-Na Tao / Jiao Qin / Qing-Tao He / Chao Zhang / Sheng-Chao Guo / Ya-Qin Du / Li-Nan Chen / Dan-Dan Shen / Zhi-Shuai Yang / Han-Qiong Zhang / Shen-Ming Huang / Yong-Hao He / Jie Cheng / Ya-Ni Zhong / Pan Shang / Jun Chen / Dao-Lai Zhang / Qian-Lang Wang / Mei-Xia Liu / Guo-Yu Li / Yongyuan Guo / H Eric Xu / Chuanxin Wang / Cheng Zhang / Shiqing Feng / Xiao Yu / Yan Zhang / Jin-Peng Sun /  Abstract: Individual free fatty acids (FAs) play important roles in metabolic homeostasis, many through engagement with more than 40G protein-coupled receptors. Searching for receptors to sense beneficial ...Individual free fatty acids (FAs) play important roles in metabolic homeostasis, many through engagement with more than 40G protein-coupled receptors. Searching for receptors to sense beneficial omega-3 FAs of fish oil enabled the identification of GPR120, which is involved in a spectrum of metabolic diseases. Here, we report six cryo-electron microscopy structures of GPR120 in complex with FA hormones or TUG891 and G or G trimers. Aromatic residues inside the GPR120 ligand pocket were responsible for recognizing different double-bond positions of these FAs and connect ligand recognition to distinct effector coupling. We also investigated synthetic ligand selectivity and the structural basis of missense single-nucleotide polymorphisms. We reveal how GPR120 differentiates rigid double bonds and flexible single bonds. The knowledge gleaned here may facilitate rational drug design targeting to GPR120. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8g59.cif.gz 8g59.cif.gz | 211.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8g59.ent.gz pdb8g59.ent.gz | 160 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8g59.json.gz 8g59.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g5/8g59 https://data.pdbj.org/pub/pdb/validation_reports/g5/8g59 ftp://data.pdbj.org/pub/pdb/validation_reports/g5/8g59 ftp://data.pdbj.org/pub/pdb/validation_reports/g5/8g59 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  29736MC  8id3C  8id4C  8id6C  8id8C  8id9C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Guanine nucleotide-binding protein ... , 3 types, 3 molecules BYA
| #1: Protein | Mass: 37285.734 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNB1 / Production host: Homo sapiens (human) / Gene: GNB1 / Production host:  |
|---|---|
| #2: Protein | Mass: 7861.143 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNG2 / Production host: Homo sapiens (human) / Gene: GNG2 / Production host:  |
| #4: Protein | Mass: 40555.199 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAI1, GNAQ, GAQ / Variant: G203A A326S / Production host: Homo sapiens (human) / Gene: GNAI1, GNAQ, GAQ / Variant: G203A A326S / Production host:  |
-Antibody / Protein / Non-polymers , 3 types, 3 molecules SR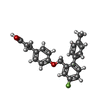

| #3: Antibody | Mass: 30363.043 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  |
|---|---|
| #5: Protein | Mass: 40533.211 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: FFAR4, GPR120, GPR129, O3FAR1, PGR4 / Production host: Homo sapiens (human) / Gene: FFAR4, GPR120, GPR129, O3FAR1, PGR4 / Production host:  |
| #6: Chemical | ChemComp-YN9 / |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-EM structure of the TUG891 bound GPR120-Giq complex Type: COMPLEX / Entity ID: #1-#5 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2000 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 52 e/Å2 / Film or detector model: FEI FALCON IV (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.64 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 303739 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj






























