[English] 日本語
 Yorodumi
Yorodumi- PDB-8et9: Cryo-EM structure of the organic cation transporter 2 in complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8et9 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the organic cation transporter 2 in complex with 1-methyl-4-phenylpyridinium | ||||||
 Components Components | OCT2 | ||||||
 Keywords Keywords | TRANSPORT PROTEIN / Cation / transport / membrane protein | ||||||
| Function / homology | MFS general substrate transporter like domains / Growth Hormone; Chain: A; / Up-down Bundle / Mainly Alpha / 1-methyl-4-phenylpyridin-1-ium Function and homology information Function and homology information | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.61 Å | ||||||
 Authors Authors | Suo, Y. / Wright, N.J. / Lee, S.-Y. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Molecular basis of polyspecific drug and xenobiotic recognition by OCT1 and OCT2. Authors: Yang Suo / Nicholas J Wright / Hugo Guterres / Justin G Fedor / Kevin John Butay / Mario J Borgnia / Wonpil Im / Seok-Yong Lee /  Abstract: A wide range of endogenous and xenobiotic organic ions require facilitated transport systems to cross the plasma membrane for their disposition. In mammals, organic cation transporter (OCT) subtypes ...A wide range of endogenous and xenobiotic organic ions require facilitated transport systems to cross the plasma membrane for their disposition. In mammals, organic cation transporter (OCT) subtypes 1 and 2 (OCT1 and OCT2, also known as SLC22A1 and SLC22A2, respectively) are polyspecific transporters responsible for the uptake and clearance of structurally diverse cationic compounds in the liver and kidneys, respectively. Notably, it is well established that human OCT1 and OCT2 play central roles in the pharmacokinetics and drug-drug interactions of many prescription medications, including metformin. Despite their importance, the basis of polyspecific cationic drug recognition and the alternating access mechanism for OCTs have remained a mystery. Here we present four cryo-electron microscopy structures of apo, substrate-bound and drug-bound OCT1 and OCT2 consensus variants, in outward-facing and outward-occluded states. Together with functional experiments, in silico docking and molecular dynamics simulations, these structures uncover general principles of organic cation recognition by OCTs and provide insights into extracellular gate occlusion. Our findings set the stage for a comprehensive structure-based understanding of OCT-mediated drug-drug interactions, which will prove critical in the preclinical evaluation of emerging therapeutics. #1: Journal: bioRxiv / Year: 2023 Title: Molecular basis of polyspecific drug binding and transport by OCT1 and OCT2. Authors: Yang Suo / Nicholas J Wright / Hugo Guterres / Justin G Fedor / Kevin John Butay / Mario J Borgnia / Wonpil Im / Seok-Yong Lee /  Abstract: A wide range of endogenous and xenobiotic organic ions require facilitated transport systems to cross the plasma membrane for their disposition . In mammals, organic cation transporter subtypes 1 ...A wide range of endogenous and xenobiotic organic ions require facilitated transport systems to cross the plasma membrane for their disposition . In mammals, organic cation transporter subtypes 1 and 2 (OCT1 and OCT2, also known as SLC22A1 and SLC22A2, respectively) are polyspecific transporters responsible for the uptake and clearance of structurally diverse cationic compounds in the liver and kidneys, respectively . Notably, it is well established that human OCT1 and OCT2 play central roles in the pharmacokinetics, pharmacodynamics, and drug-drug interactions (DDI) of many prescription medications, including metformin . Despite their importance, the basis of polyspecific cationic drug recognition and the alternating access mechanism for OCTs have remained a mystery. Here, we present four cryo-EM structures of apo, substrate-bound, and drug-bound OCT1 and OCT2 in outward-facing and outward-occluded states. Together with functional experiments, docking, and molecular dynamics simulations, these structures uncover general principles of organic cation recognition by OCTs and illuminate unexpected features of the OCT alternating access mechanism. Our findings set the stage for a comprehensive structure-based understanding of OCT-mediated DDI, which will prove critical in the preclinical evaluation of emerging therapeutics. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8et9.cif.gz 8et9.cif.gz | 171.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8et9.ent.gz pdb8et9.ent.gz | 134 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8et9.json.gz 8et9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/et/8et9 https://data.pdbj.org/pub/pdb/validation_reports/et/8et9 ftp://data.pdbj.org/pub/pdb/validation_reports/et/8et9 ftp://data.pdbj.org/pub/pdb/validation_reports/et/8et9 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  28589MC  8et6C  8et7C  8et8C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 62030.547 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:  Homo sapiens (human) Homo sapiens (human) |
|---|---|
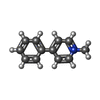
| #2: Chemical | ChemComp-WRF / |
| Has ligand of interest | Y |
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: OCT2 / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 60 kDa/nm / Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||
| Buffer solution | pH: 8 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R1.2/1.3 | ||||||||||||||||||||
| Vitrification | Instrument: LEICA EM GP / Cryogen name: ETHANE / Humidity: 90 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 81000 X / Nominal defocus max: 2000 nm / Nominal defocus min: 800 nm / Cs: 2.7 mm / C2 aperture diameter: 50 µm / Alignment procedure: ZEMLIN TABLEAU |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 3.7 sec. / Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 8029 |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
| Image scans | Width: 5760 / Height: 4092 |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1141906 | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.61 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 73474 / Algorithm: FOURIER SPACE / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | B value: 141.9 / Protocol: AB INITIO MODEL / Space: REAL | ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj