[English] 日本語
 Yorodumi
Yorodumi- PDB-8ar0: Solution structure of TLR2 transmembrane and cytoplasmic juxtamem... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8ar0 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Solution structure of TLR2 transmembrane and cytoplasmic juxtamembrane regions | ||||||
 Components Components | Toll-like receptor 2 | ||||||
 Keywords Keywords | MEMBRANE PROTEIN / PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationToll Like Receptor TLR6:TLR2 Cascade / triacyl lipopeptide binding / Toll-like receptor 2-Toll-like receptor 6 protein complex / detection of diacyl bacterial lipopeptide / cellular response to diacyl bacterial lipopeptide / Toll-like receptor 1-Toll-like receptor 2 protein complex / detection of triacyl bacterial lipopeptide / cellular response to triacyl bacterial lipopeptide / cellular response to bacterial lipopeptide / negative regulation of synapse assembly ...Toll Like Receptor TLR6:TLR2 Cascade / triacyl lipopeptide binding / Toll-like receptor 2-Toll-like receptor 6 protein complex / detection of diacyl bacterial lipopeptide / cellular response to diacyl bacterial lipopeptide / Toll-like receptor 1-Toll-like receptor 2 protein complex / detection of triacyl bacterial lipopeptide / cellular response to triacyl bacterial lipopeptide / cellular response to bacterial lipopeptide / negative regulation of synapse assembly / positive regulation of cellular response to macrophage colony-stimulating factor stimulus / Toll Like Receptor TLR1:TLR2 Cascade / Beta defensins / lipopolysaccharide immune receptor activity / toll-like receptor 2 signaling pathway / positive regulation of matrix metallopeptidase secretion / Toll-like receptor binding / toll-like receptor TLR6:TLR2 signaling pathway / positive regulation of interleukin-18 production / Regulation of TLR by endogenous ligand / peptidoglycan binding / NAD+ nucleosidase activity, cyclic ADP-ribose generating / MyD88 deficiency (TLR2/4) / microglia development / IRAK4 deficiency (TLR2/4) / MyD88:MAL(TIRAP) cascade initiated on plasma membrane / toll-like receptor signaling pathway / negative regulation of phagocytosis / pattern recognition receptor activity / RSV-host interactions / cellular response to lipoteichoic acid / positive regulation of Wnt signaling pathway / positive regulation of chemokine production / positive regulation of interleukin-12 production / positive regulation of interferon-beta production / secretory granule membrane / learning / positive regulation of interleukin-8 production / lipopolysaccharide binding / : / positive regulation of interleukin-6 production / cellular response to type II interferon / phagocytic vesicle membrane / positive regulation of tumor necrosis factor production / positive regulation of inflammatory response / Modulation by Mtb of host immune system / transmembrane signaling receptor activity / signaling receptor activity / amyloid-beta binding / ER-Phagosome pathway / defense response to virus / positive regulation of canonical NF-kappaB signal transduction / receptor complex / defense response to Gram-positive bacterium / immune response / membrane raft / inflammatory response / intracellular membrane-bounded organelle / innate immune response / apoptotic process / Neutrophil degranulation / positive regulation of gene expression / protein-containing complex binding / SARS-CoV-2 activates/modulates innate and adaptive immune responses / cell surface / Golgi apparatus / signal transduction / positive regulation of transcription by RNA polymerase II / nucleoplasm / identical protein binding / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | SOLUTION NMR / torsion angle dynamics | ||||||
 Authors Authors | Kornilov, F.D. / Shabalkina, A.V. / Goncharuk, M.V. / Goncharuk, S.A. / Arseniev, A.S. / Mineev, K.S. | ||||||
| Funding support |  Russian Federation, 1items Russian Federation, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors. Authors: Kornilov, F.D. / Shabalkina, A.V. / Lin, C. / Volynsky, P.E. / Kot, E.F. / Kayushin, A.L. / Lushpa, V.A. / Goncharuk, M.V. / Arseniev, A.S. / Goncharuk, S.A. / Wang, X. / Mineev, K.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8ar0.cif.gz 8ar0.cif.gz | 170.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8ar0.ent.gz pdb8ar0.ent.gz | 139.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8ar0.json.gz 8ar0.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ar/8ar0 https://data.pdbj.org/pub/pdb/validation_reports/ar/8ar0 ftp://data.pdbj.org/pub/pdb/validation_reports/ar/8ar0 ftp://data.pdbj.org/pub/pdb/validation_reports/ar/8ar0 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  8ar1C  8ar2C  8ar3C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 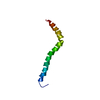
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 5852.218 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TLR2, TIL4 / Production host: Homo sapiens (human) / Gene: TLR2, TIL4 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj







 HSQC
HSQC