+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7zfu | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BRD4 in complex with PepLite-Pro | ||||||||||||
 Components Components | Bromodomain-containing protein 4 | ||||||||||||
 Keywords Keywords | TRANSCRIPTION / FragLites / anomalous / BRD4 / Fragment screening | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase II C-terminal domain binding / P-TEFb complex binding / negative regulation of DNA damage checkpoint / histone H4 reader activity / host-mediated suppression of viral transcription / positive regulation of G2/M transition of mitotic cell cycle / positive regulation of T-helper 17 cell lineage commitment / : / RNA polymerase II CTD heptapeptide repeat kinase activity / condensed nuclear chromosome ...RNA polymerase II C-terminal domain binding / P-TEFb complex binding / negative regulation of DNA damage checkpoint / histone H4 reader activity / host-mediated suppression of viral transcription / positive regulation of G2/M transition of mitotic cell cycle / positive regulation of T-helper 17 cell lineage commitment / : / RNA polymerase II CTD heptapeptide repeat kinase activity / condensed nuclear chromosome / transcription coregulator activity / positive regulation of transcription elongation by RNA polymerase II / p53 binding / chromosome / regulation of inflammatory response / histone binding / Potential therapeutics for SARS / transcription coactivator activity / positive regulation of canonical NF-kappaB signal transduction / transcription cis-regulatory region binding / chromatin remodeling / protein serine/threonine kinase activity / DNA damage response / chromatin binding / regulation of transcription by RNA polymerase II / chromatin / positive regulation of DNA-templated transcription / enzyme binding / positive regulation of transcription by RNA polymerase II / nucleoplasm / nucleus Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.29 Å MOLECULAR REPLACEMENT / Resolution: 1.29 Å | ||||||||||||
 Authors Authors | Turberville, S. / Martin, M.P. / Hope, I. / Noble, M.E.M. | ||||||||||||
| Funding support |  United Kingdom, 3items United Kingdom, 3items
| ||||||||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2022 Journal: J.Med.Chem. / Year: 2022Title: Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions. Authors: Davison, G. / Martin, M.P. / Turberville, S. / Dormen, S. / Heath, R. / Heptinstall, A.B. / Lawson, M. / Miller, D.C. / Ng, Y.M. / Sanderson, J.N. / Hope, I. / Wood, D.J. / Cano, C. / ...Authors: Davison, G. / Martin, M.P. / Turberville, S. / Dormen, S. / Heath, R. / Heptinstall, A.B. / Lawson, M. / Miller, D.C. / Ng, Y.M. / Sanderson, J.N. / Hope, I. / Wood, D.J. / Cano, C. / Endicott, J.A. / Hardcastle, I.R. / Noble, M.E.M. / Waring, M.J. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7zfu.cif.gz 7zfu.cif.gz | 47.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7zfu.ent.gz pdb7zfu.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7zfu.json.gz 7zfu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7zfu_validation.pdf.gz 7zfu_validation.pdf.gz | 732 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7zfu_full_validation.pdf.gz 7zfu_full_validation.pdf.gz | 733.2 KB | Display | |
| Data in XML |  7zfu_validation.xml.gz 7zfu_validation.xml.gz | 9.2 KB | Display | |
| Data in CIF |  7zfu_validation.cif.gz 7zfu_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zf/7zfu https://data.pdbj.org/pub/pdb/validation_reports/zf/7zfu ftp://data.pdbj.org/pub/pdb/validation_reports/zf/7zfu ftp://data.pdbj.org/pub/pdb/validation_reports/zf/7zfu | HTTPS FTP |
-Related structure data
| Related structure data |  7ppxC  7qu7C  7qukC  7qumC  7qwoC  7qx1C  7qxtC  7qykC  7qylC  7qzmC  7qzyC  7qzzC  7r00C  7r05C  7r0yC  7z9hC  7z9iC  7z9jC  7z9nC  7z9oC  7z9sC  7z9uC  7z9wC  7z9yC  7za6C  7za7C  7za8C  7za9C  7zaaC  7zadC  7zaeC  7zajC  7zaqC  7zarC  7zatC  7ze6C  7ze7C  7zefC  7zenC  7zfnC  7zfoC  7zfsC  7zftC  7zfvC  7zfyC  7zfzC  7zg1C  7zg2C  5lrqS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 15294.578 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: BRD4, HUNK1 / Plasmid: pET28b / Production host: Homo sapiens (human) / Gene: BRD4, HUNK1 / Plasmid: pET28b / Production host:  | ||||
|---|---|---|---|---|---|
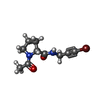
| #2: Chemical | ChemComp-IS6 / ( | ||||
| #3: Chemical | | #4: Water | ChemComp-HOH / | Has ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.16 Å3/Da / Density % sol: 43.05 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8.5 Details: 0.1 M Buffer system 3 (1 M Tris, 1 M BICINE, pH 8.5), 34-44% Precipitant Solution 3 (20% PEG 4000, 40%) and 60-80 mM Halogens Mix (NaF, NaBr and NaI) solution |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04-1 / Wavelength: 0.97623 Å / Beamline: I04-1 / Wavelength: 0.97623 Å | |||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Jul 28, 2019 | |||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97623 Å / Relative weight: 1 | |||||||||||||||||||||
| Reflection | Resolution: 1.29→38.19 Å / Num. obs: 33926 / % possible obs: 99.3 % / Redundancy: 7.1 % / CC1/2: 0.988 / Rmerge(I) obs: 0.097 / Rpim(I) all: 0.056 / Rrim(I) all: 0.112 / Net I/σ(I): 8.5 | |||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5LRQ Resolution: 1.29→38.19 Å / Cor.coef. Fo:Fc: 0.965 / Cor.coef. Fo:Fc free: 0.965 / SU B: 0.992 / SU ML: 0.04 / Cross valid method: FREE R-VALUE / ESU R: 0.053 / ESU R Free: 0.052 Details: Hydrogens have been added in their riding positions
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20.689 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.29→38.19 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj