+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7z9s | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | ATAD2 in complex with PepLite-Arg | ||||||||||||
 Components Components | ATPase family AAA domain-containing protein 2 | ||||||||||||
 Keywords Keywords | TRANSCRIPTION / ATAD2 / INHIBITOR / FRAGMENT / BROMODOMAIN / FRAGLITE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnucleosome disassembly / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / transcription initiation-coupled chromatin remodeling / nucleosome assembly / histone binding / chromatin binding / positive regulation of DNA-templated transcription / ATP hydrolysis activity / extracellular exosome ...nucleosome disassembly / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / TFAP2 (AP-2) family regulates transcription of growth factors and their receptors / transcription initiation-coupled chromatin remodeling / nucleosome assembly / histone binding / chromatin binding / positive regulation of DNA-templated transcription / ATP hydrolysis activity / extracellular exosome / nucleoplasm / ATP binding / nucleus Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å MOLECULAR REPLACEMENT / Resolution: 1.5 Å | ||||||||||||
 Authors Authors | Turberville, S. / Martin, M.P. / Hope, I. / Noble, M.E.M. | ||||||||||||
| Funding support |  United Kingdom, 3items United Kingdom, 3items
| ||||||||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2022 Journal: J.Med.Chem. / Year: 2022Title: Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions. Authors: Davison, G. / Martin, M.P. / Turberville, S. / Dormen, S. / Heath, R. / Heptinstall, A.B. / Lawson, M. / Miller, D.C. / Ng, Y.M. / Sanderson, J.N. / Hope, I. / Wood, D.J. / Cano, C. / ...Authors: Davison, G. / Martin, M.P. / Turberville, S. / Dormen, S. / Heath, R. / Heptinstall, A.B. / Lawson, M. / Miller, D.C. / Ng, Y.M. / Sanderson, J.N. / Hope, I. / Wood, D.J. / Cano, C. / Endicott, J.A. / Hardcastle, I.R. / Noble, M.E.M. / Waring, M.J. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7z9s.cif.gz 7z9s.cif.gz | 51.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7z9s.ent.gz pdb7z9s.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7z9s.json.gz 7z9s.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/z9/7z9s https://data.pdbj.org/pub/pdb/validation_reports/z9/7z9s ftp://data.pdbj.org/pub/pdb/validation_reports/z9/7z9s ftp://data.pdbj.org/pub/pdb/validation_reports/z9/7z9s | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7ppxC  7qu7C  7qukC  7qumC  7qwoC  7qx1C  7qxtC  7qykC  7qylC  7qzmC  7qzyC  7qzzC  7r00C  7r05C  7r0yC  7z9hC  7z9iC  7z9jC  7z9nC  7z9oC  7z9uC  7z9wC  7z9yC  7za6C  7za7C  7za8C  7za9C  7zaaC  7zadC  7zaeC  7zajC  7zaqC  7zarC  7zatC  7ze6C  7ze7C  7zefC  7zenC  7zfnC  7zfoC  7zfsC  7zftC  7zfuC  7zfvC  7zfyC  7zfzC  7zg1C  7zg2C  3daiS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules AAA
| #1: Protein | Mass: 15453.514 Da / Num. of mol.: 1 / Fragment: bromodomain Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ATAD2, L16, PRO2000 / Plasmid: pET28b / Production host: Homo sapiens (human) / Gene: ATAD2, L16, PRO2000 / Plasmid: pET28b / Production host:  |
|---|
-Non-polymers , 5 types, 207 molecules 

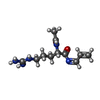






| #2: Chemical | | #3: Chemical | ChemComp-CL / | #4: Chemical | ChemComp-IJI / ( | #5: Chemical | ChemComp-EDO / #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | N |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6.5 / Details: 0.1M BisTris pH 6-7, 1.7-2.1M Ammonium sulphate / PH range: 6-7 |
|---|
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04-1 / Wavelength: 0.91162 Å / Beamline: I04-1 / Wavelength: 0.91162 Å | |||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Sep 25, 2019 | |||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.91162 Å / Relative weight: 1 | |||||||||||||||||||||
| Reflection | Resolution: 1.5→48.57 Å / Num. obs: 41696 / % possible obs: 100 % / Redundancy: 39.6 % / CC1/2: 1 / Rmerge(I) obs: 0.115 / Rpim(I) all: 0.025 / Rrim(I) all: 0.118 / Net I/σ(I): 19 | |||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3DAI Resolution: 1.5→48.57 Å / Cor.coef. Fo:Fc: 0.967 / Cor.coef. Fo:Fc free: 0.96 / SU B: 1.504 / SU ML: 0.053 / Cross valid method: FREE R-VALUE / ESU R: 0.062 / ESU R Free: 0.067 Details: Hydrogens have been added in their riding positions
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 35.981 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.5→48.57 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj









