[English] 日本語
 Yorodumi
Yorodumi- PDB-7xr6: Structure of human excitatory amino acid transporter 2 (EAAT2) in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7xr6 | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
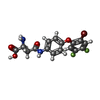
| Title | Structure of human excitatory amino acid transporter 2 (EAAT2) in complex with WAY-213613 | |||||||||||||||||||||||||||||||||
 Components Components | Excitatory amino acid transporter 2 | |||||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSPORT PROTEIN / Glutamate transport | |||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationneurotransmitter reuptake / Astrocytic Glutamate-Glutamine Uptake And Metabolism / cysteine transmembrane transporter activity / high-affinity L-glutamate transmembrane transporter activity / glutamate:sodium symporter activity / visual behavior / membrane protein complex / : / L-glutamate transmembrane transporter activity / L-glutamate transmembrane transport ...neurotransmitter reuptake / Astrocytic Glutamate-Glutamine Uptake And Metabolism / cysteine transmembrane transporter activity / high-affinity L-glutamate transmembrane transporter activity / glutamate:sodium symporter activity / visual behavior / membrane protein complex / : / L-glutamate transmembrane transporter activity / L-glutamate transmembrane transport / glutathione biosynthetic process / D-aspartate import across plasma membrane / L-aspartate transmembrane transport / monoatomic anion transmembrane transporter activity / telencephalon development / L-aspartate import across plasma membrane / Glutamate Neurotransmitter Release Cycle / L-glutamate import across plasma membrane / neutral L-amino acid transmembrane transporter activity / transepithelial transport / neuron projection terminus / cellular response to cocaine / astrocyte projection / neurotransmitter transport / protein homotrimerization / adult behavior / axolemma / response to amino acid / transport across blood-brain barrier / monoatomic ion transport / positive regulation of D-glucose import across plasma membrane / response to wounding / multicellular organism growth / presynaptic membrane / cell body / vesicle / chemical synaptic transmission / membrane raft / response to xenobiotic stimulus / glutamatergic synapse / cell surface / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Zhao, Y. / Zhang, Z. | |||||||||||||||||||||||||||||||||
| Funding support |  China, 1items China, 1items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of ligand binding modes of human EAAT2. Authors: Zhenglai Zhang / Huiwen Chen / Ze Geng / Zhuoya Yu / Hang Li / Yanli Dong / Hongwei Zhang / Zhuo Huang / Juquan Jiang / Yan Zhao /  Abstract: In the central nervous system (CNS), excitatory amino acid transporters (EAATs) mediate the uptake of excitatory neurotransmitter glutamate and maintain its low concentrations in the synaptic cleft ...In the central nervous system (CNS), excitatory amino acid transporters (EAATs) mediate the uptake of excitatory neurotransmitter glutamate and maintain its low concentrations in the synaptic cleft for avoiding neuronal cytotoxicity. Dysfunction of EAATs can lead to many psychiatric diseases. Here we report cryo-EM structures of human EAAT2 in an inward-facing conformation, in the presence of substrate glutamate or selective inhibitor WAY-213613. The glutamate is coordinated by extensive hydrogen bonds and further stabilized by HP2. The inhibitor WAY-213613 occupies a similar binding pocket to that of the substrate glutamate. Upon association with the WAY-213613, the HP2 undergoes a substantial conformational change, and in turn stabilizes the inhibitor binding by forming hydrophobic interactions. Electrophysiological experiments elucidate that the unique S441 plays pivotal roles in the binding of hEAAT2 with glutamate or WAY-213613, and the I464-L467-V468 cluster acts as a key structural determinant for the selective inhibition of this transporter by WAY-213613. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7xr6.cif.gz 7xr6.cif.gz | 245.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7xr6.ent.gz pdb7xr6.ent.gz | 189.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7xr6.json.gz 7xr6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xr/7xr6 https://data.pdbj.org/pub/pdb/validation_reports/xr/7xr6 ftp://data.pdbj.org/pub/pdb/validation_reports/xr/7xr6 ftp://data.pdbj.org/pub/pdb/validation_reports/xr/7xr6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  33408MC  7xr4C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 62164.977 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SLC1A2, EAAT2, GLT1 / Production host: Homo sapiens (human) / Gene: SLC1A2, EAAT2, GLT1 / Production host:  Homo sapiens (human) / References: UniProt: P43004 Homo sapiens (human) / References: UniProt: P43004#2: Chemical | #3: Chemical | #4: Chemical | ChemComp-PC1 / #5: Sugar | Has ligand of interest | Y | Has protein modification | N | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: transporter / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 2500 nm / Nominal defocus min: 1200 nm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 420 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj