[English] 日本語
 Yorodumi
Yorodumi- EMDB-33408: Structure of human excitatory amino acid transporter 2 (EAAT2) in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human excitatory amino acid transporter 2 (EAAT2) in complex with WAY-213613 | |||||||||
 Map data Map data | EAAT2_WAY213.mrc | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glutamate transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationneurotransmitter reuptake / Astrocytic Glutamate-Glutamine Uptake And Metabolism / cysteine transmembrane transporter activity / high-affinity L-glutamate transmembrane transporter activity / glutamate:sodium symporter activity / visual behavior / membrane protein complex / : / L-glutamate transmembrane transporter activity / L-glutamate transmembrane transport ...neurotransmitter reuptake / Astrocytic Glutamate-Glutamine Uptake And Metabolism / cysteine transmembrane transporter activity / high-affinity L-glutamate transmembrane transporter activity / glutamate:sodium symporter activity / visual behavior / membrane protein complex / : / L-glutamate transmembrane transporter activity / L-glutamate transmembrane transport / glutathione biosynthetic process / D-aspartate import across plasma membrane / L-aspartate transmembrane transport / monoatomic anion transmembrane transporter activity / telencephalon development / L-aspartate import across plasma membrane / Glutamate Neurotransmitter Release Cycle / L-glutamate import across plasma membrane / neutral L-amino acid transmembrane transporter activity / transepithelial transport / neuron projection terminus / cellular response to cocaine / astrocyte projection / neurotransmitter transport / protein homotrimerization / adult behavior / axolemma / response to amino acid / transport across blood-brain barrier / monoatomic ion transport / positive regulation of D-glucose import across plasma membrane / response to wounding / multicellular organism growth / presynaptic membrane / cell body / vesicle / chemical synaptic transmission / membrane raft / response to xenobiotic stimulus / glutamatergic synapse / cell surface / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Yan Z / Zhenglai Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis of ligand binding modes of human EAAT2. Authors: Zhenglai Zhang / Huiwen Chen / Ze Geng / Zhuoya Yu / Hang Li / Yanli Dong / Hongwei Zhang / Zhuo Huang / Juquan Jiang / Yan Zhao /  Abstract: In the central nervous system (CNS), excitatory amino acid transporters (EAATs) mediate the uptake of excitatory neurotransmitter glutamate and maintain its low concentrations in the synaptic cleft ...In the central nervous system (CNS), excitatory amino acid transporters (EAATs) mediate the uptake of excitatory neurotransmitter glutamate and maintain its low concentrations in the synaptic cleft for avoiding neuronal cytotoxicity. Dysfunction of EAATs can lead to many psychiatric diseases. Here we report cryo-EM structures of human EAAT2 in an inward-facing conformation, in the presence of substrate glutamate or selective inhibitor WAY-213613. The glutamate is coordinated by extensive hydrogen bonds and further stabilized by HP2. The inhibitor WAY-213613 occupies a similar binding pocket to that of the substrate glutamate. Upon association with the WAY-213613, the HP2 undergoes a substantial conformational change, and in turn stabilizes the inhibitor binding by forming hydrophobic interactions. Electrophysiological experiments elucidate that the unique S441 plays pivotal roles in the binding of hEAAT2 with glutamate or WAY-213613, and the I464-L467-V468 cluster acts as a key structural determinant for the selective inhibition of this transporter by WAY-213613. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33408.map.gz emd_33408.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33408-v30.xml emd-33408-v30.xml emd-33408.xml emd-33408.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33408.png emd_33408.png | 64.3 KB | ||
| Filedesc metadata |  emd-33408.cif.gz emd-33408.cif.gz | 6.2 KB | ||
| Others |  emd_33408_half_map_1.map.gz emd_33408_half_map_1.map.gz emd_33408_half_map_2.map.gz emd_33408_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33408 http://ftp.pdbj.org/pub/emdb/structures/EMD-33408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33408 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33408 | HTTPS FTP |
-Related structure data
| Related structure data |  7xr6MC  7xr4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33408.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33408.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EAAT2_WAY213.mrc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: map half A.mrc
| File | emd_33408_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map_half_A.mrc | ||||||||||||
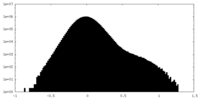
| Projections & Slices |
| ||||||||||||
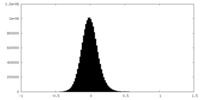
| Density Histograms |
-Half map: map half B.mrc
| File | emd_33408_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map_half_B.mrc | ||||||||||||
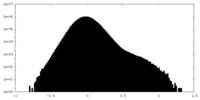
| Projections & Slices |
| ||||||||||||
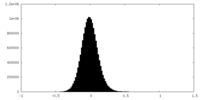
| Density Histograms |
- Sample components
Sample components
-Entire : transporter
| Entire | Name: transporter |
|---|---|
| Components |
|
-Supramolecule #1: transporter
| Supramolecule | Name: transporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Excitatory amino acid transporter 2
| Macromolecule | Name: Excitatory amino acid transporter 2 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 62.164977 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASTEGANNM PKQVEVRMHD SHLGSEEPKH RHLGLRLCDK LGKNLLLTLT VFGVILGAVC GGLLRLASPI HPDVVMLIAF PGDILMRML KMLILPLIIS SLITGLSGLD AKASGRLGTR AMVYYMSTTI IAAVLGVILV LAIHPGNPKL KKQLGPGKKN D EVSSLDAF ...String: MASTEGANNM PKQVEVRMHD SHLGSEEPKH RHLGLRLCDK LGKNLLLTLT VFGVILGAVC GGLLRLASPI HPDVVMLIAF PGDILMRML KMLILPLIIS SLITGLSGLD AKASGRLGTR AMVYYMSTTI IAAVLGVILV LAIHPGNPKL KKQLGPGKKN D EVSSLDAF LDLIRNLFPE NLVQACFQQI QTVTKKVLVA PPPDEEANAT SAVVSLLNET VTEVPEETKM VIKKGLEFKD GM NVLGLIG FFIAFGIAMG KMGDQAKLMV DFFNILNEIV MKLVIMIMWY SPLGIACLIC GKIIAIKDLE VVARQLGMYM VTV IIGLII HGGIFLPLIY FVVTRKNPFS FFAGIFQAWI TALGTASSAG TLPVTFRCLE ENLGIDKRVT RFVLPVGATI NMDG TALYE AVAAIFIAQM NGVVLDGGQI VTVSLTATLA SVGAASIPSA GLVTMLLILT AVGLPTEDIS LLVAVDWLLD RMRTS VNVV GDSFGAGIVY HLSKSELDTI DSQHRVHEDI EMTKTQSIYD DMKNHRESNS NQCVYAAHNS VIVDECKVTL AANGKS ADC SVEEEPWKRE K UniProtKB: Excitatory amino acid transporter 2 |
-Macromolecule #2: (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]...
| Macromolecule | Name: (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid type: ligand / ID: 2 / Number of copies: 3 / Formula: GJ0 |
|---|---|
| Molecular weight | Theoretical: 415.186 Da |
| Chemical component information | 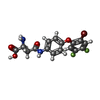 ChemComp-GJ0: |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 3 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 4 / Number of copies: 30 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #5: dodecyl beta-D-glucopyranoside
| Macromolecule | Name: dodecyl beta-D-glucopyranoside / type: ligand / ID: 5 / Number of copies: 3 / Formula: XKJ |
|---|---|
| Molecular weight | Theoretical: 348.475 Da |
| Chemical component information | 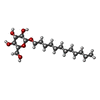 ChemComp-XKJ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)