+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7wnh | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Nurr1 binding to NBRE | ||||||
 Components Components |
| ||||||
 Keywords Keywords | TRANSCRIPTION / Nurr1 / NBRE / DBD / LBD | ||||||
| Function / homology |  Function and homology information Function and homology informationgeneral adaptation syndrome / habenula development / cellular response to corticotropin-releasing hormone stimulus / central nervous system projection neuron axonogenesis / nuclear glucocorticoid receptor binding / regulation of dopamine metabolic process / midbrain dopaminergic neuron differentiation / regulation of respiratory gaseous exchange / dopaminergic neuron differentiation / neuron maturation ...general adaptation syndrome / habenula development / cellular response to corticotropin-releasing hormone stimulus / central nervous system projection neuron axonogenesis / nuclear glucocorticoid receptor binding / regulation of dopamine metabolic process / midbrain dopaminergic neuron differentiation / regulation of respiratory gaseous exchange / dopaminergic neuron differentiation / neuron maturation / central nervous system neuron differentiation / dopamine biosynthetic process / fat cell differentiation / negative regulation of apoptotic signaling pathway / canonical Wnt signaling pathway / nuclear retinoid X receptor binding / response to amphetamine / adult locomotory behavior / post-embryonic development / SUMOylation of intracellular receptors / beta-catenin binding / Nuclear Receptor transcription pathway / nuclear receptor activity / neuron migration / sequence-specific double-stranded DNA binding / cellular response to oxidative stress / DNA-binding transcription activator activity, RNA polymerase II-specific / neuron apoptotic process / transcription regulator complex / negative regulation of neuron apoptotic process / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific / response to hypoxia / nuclear speck / RNA polymerase II cis-regulatory region sequence-specific DNA binding / protein heterodimerization activity / DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / protein-containing complex / DNA binding / zinc ion binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.1 Å MOLECULAR REPLACEMENT / Resolution: 3.1 Å | ||||||
 Authors Authors | Zhao, M. / Xu, T. / Wang, N. / Guo, Y. / Liu, J. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2022 Journal: Proc.Natl.Acad.Sci.USA / Year: 2022Title: Integrative analysis reveals structural basis for transcription activation of Nurr1 and Nurr1-RXR alpha heterodimer. Authors: Zhao, M. / Wang, N. / Guo, Y. / Li, J. / Yin, Y. / Dong, Y. / Zhu, J. / Peng, C. / Xu, T. / Liu, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7wnh.cif.gz 7wnh.cif.gz | 330.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7wnh.ent.gz pdb7wnh.ent.gz | 259.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7wnh.json.gz 7wnh.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wn/7wnh https://data.pdbj.org/pub/pdb/validation_reports/wn/7wnh ftp://data.pdbj.org/pub/pdb/validation_reports/wn/7wnh ftp://data.pdbj.org/pub/pdb/validation_reports/wn/7wnh | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 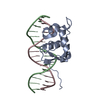 1citS  1ovlS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: _ / Refine code: _
|
 Movie
Movie Controller
Controller



 PDBj
PDBj





































