Entry Database : PDB / ID : 7wkuTitle Structure of PDCoV Mpro in complex with an inhibitor N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE Peptidase C30 Keywords / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species synthetic construct (others) Method / / / Resolution : 2.6 Å Authors Wang, F.H. / Yang, H.T. Funding support Organization Grant number Country National Natural Science Foundation of China (NSFC) 81772204
Journal : Viruses / Year : 2022Title : The Structure of the Porcine Deltacoronavirus Main Protease Reveals a Conserved Target for the Design of Antivirals.Authors : Wang, F. / Chen, C. / Wang, Z. / Han, X. / Shi, P. / Zhou, K. / Liu, X. / Xiao, Y. / Cai, Y. / Huang, J. / Zhang, L. / Yang, H. History Deposition Jan 11, 2022 Deposition site / Processing site Revision 1.0 May 25, 2022 Provider / Type Revision 2.0 Nov 15, 2023 Group / Data collection / Derived calculationsCategory atom_site / chem_comp_atom ... atom_site / chem_comp_atom / chem_comp_bond / pdbx_validate_rmsd_bond / pdbx_validate_torsion / struct_conn Item _atom_site.auth_atom_id / _atom_site.label_atom_id ... _atom_site.auth_atom_id / _atom_site.label_atom_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr2_label_atom_id Revision 2.1 Nov 29, 2023 Group / Category Revision 2.2 Oct 16, 2024 Group / Category / pdbx_modification_feature / Item
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Porcine deltacoronavirus
Porcine deltacoronavirus X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å
MOLECULAR REPLACEMENT / Resolution: 2.6 Å  Authors
Authors China, 1items
China, 1items  Citation
Citation Journal: Viruses / Year: 2022
Journal: Viruses / Year: 2022 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7wku.cif.gz
7wku.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7wku.ent.gz
pdb7wku.ent.gz PDB format
PDB format 7wku.json.gz
7wku.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/wk/7wku
https://data.pdbj.org/pub/pdb/validation_reports/wk/7wku ftp://data.pdbj.org/pub/pdb/validation_reports/wk/7wku
ftp://data.pdbj.org/pub/pdb/validation_reports/wk/7wku
 F&H Search
F&H Search Links
Links Assembly
Assembly
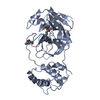





 Components
Components Porcine deltacoronavirus / Gene: ORF1ab / Production host:
Porcine deltacoronavirus / Gene: ORF1ab / Production host: 
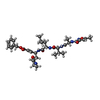
 Type: Peptide-like / Class: Inhibitor / Mass: 680.791 Da / Num. of mol.: 6 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others)
Type: Peptide-like / Class: Inhibitor / Mass: 680.791 Da / Num. of mol.: 6 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  SSRF
SSRF  / Beamline: BL19U1 / Wavelength: 0.97923 Å
/ Beamline: BL19U1 / Wavelength: 0.97923 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller



 PDBj
PDBj


