[English] 日本語
 Yorodumi
Yorodumi- PDB-7vfw: Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-b... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7vfw | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to CaV2.2-blocker1 | |||||||||||||||||||||||||||
 Components Components | (Voltage-dependent ...) x 3 | |||||||||||||||||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / Voltage gated calcium channel / N-type / complex | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationPresynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / positive regulation of calcium ion-dependent exocytosis of neurotransmitter / calcium ion transmembrane transport via high voltage-gated calcium channel / Phase 2 - plateau phase / high voltage-gated calcium channel activity / membrane depolarization during bundle of His cell action potential / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / NCAM1 interactions ...Presynaptic depolarization and calcium channel opening / regulation of membrane repolarization during action potential / positive regulation of calcium ion-dependent exocytosis of neurotransmitter / calcium ion transmembrane transport via high voltage-gated calcium channel / Phase 2 - plateau phase / high voltage-gated calcium channel activity / membrane depolarization during bundle of His cell action potential / L-type voltage-gated calcium channel complex / positive regulation of muscle contraction / NCAM1 interactions / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / calcium ion transport into cytosol / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / voltage-gated calcium channel complex / Mechanical load activates signaling by PIEZO1 and integrins in osteocytes / Phase 0 - rapid depolarisation / response to amyloid-beta / regulation of heart rate by cardiac conduction / calcium ion import across plasma membrane / regulation of calcium ion transport / neuronal dense core vesicle / voltage-gated calcium channel activity / presynaptic active zone membrane / T-tubule / sarcoplasmic reticulum / calcium channel regulator activity / modulation of chemical synaptic transmission / calcium ion transmembrane transport / GABA-ergic synapse / cellular response to amyloid-beta / calcium ion transport / amyloid-beta binding / chemical synaptic transmission / neuronal cell body / calcium ion binding / synapse / extracellular exosome / ATP binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||||||||||||||
 Authors Authors | Dong, Y. / Gao, Y. / Wang, Y. / Zhao, Y. | |||||||||||||||||||||||||||
| Funding support |  China, 1items China, 1items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2021 Journal: Cell Rep / Year: 2021Title: Closed-state inactivation and pore-blocker modulation mechanisms of human Ca2.2. Authors: Yanli Dong / Yiwei Gao / Shuai Xu / Yuhang Wang / Zhuoya Yu / Yue Li / Bin Li / Tian Yuan / Bei Yang / Xuejun Cai Zhang / Daohua Jiang / Zhuo Huang / Yan Zhao /  Abstract: N-type voltage-gated calcium (Ca) channels mediate Ca influx at presynaptic terminals in response to action potentials and play vital roles in synaptogenesis, release of neurotransmitters, and ...N-type voltage-gated calcium (Ca) channels mediate Ca influx at presynaptic terminals in response to action potentials and play vital roles in synaptogenesis, release of neurotransmitters, and nociceptive transmission. Here, we elucidate a cryo-electron microscopy (cryo-EM) structure of the human Ca2.2 complex in apo, ziconotide-bound, and two Ca2.2-specific pore blockers-bound states. The second voltage-sensing domain (VSD) is captured in a resting-state conformation, trapped by a phosphatidylinositol 4,5-bisphosphate (PIP) molecule, which is distinct from the other three VSDs of Ca2.2, as well as activated VSDs observed in previous structures of Ca channels. This structure reveals the molecular basis for the unique inactivation process of Ca2.2 channels, in which the intracellular gate formed by S6 helices is closed and a W-helix from the domain II-III linker stabilizes closed-state inactivation. The structures of this inactivated, drug-bound complex lay a solid foundation for developing new state-dependent blockers for treatment of chronic pain. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7vfw.cif.gz 7vfw.cif.gz | 484.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7vfw.ent.gz pdb7vfw.ent.gz | 370.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7vfw.json.gz 7vfw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7vfw_validation.pdf.gz 7vfw_validation.pdf.gz | 1.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7vfw_full_validation.pdf.gz 7vfw_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  7vfw_validation.xml.gz 7vfw_validation.xml.gz | 71.4 KB | Display | |
| Data in CIF |  7vfw_validation.cif.gz 7vfw_validation.cif.gz | 108.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vf/7vfw https://data.pdbj.org/pub/pdb/validation_reports/vf/7vfw ftp://data.pdbj.org/pub/pdb/validation_reports/vf/7vfw ftp://data.pdbj.org/pub/pdb/validation_reports/vf/7vfw | HTTPS FTP |
-Related structure data
| Related structure data |  31961MC  7vfsC  7vfuC  7vfvC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Voltage-dependent ... , 3 types, 3 molecules ABD
| #1: Protein | Mass: 262831.781 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CACNA1B, CACH5, CACNL1A5 / Production host: Homo sapiens (human) / Gene: CACNA1B, CACH5, CACNL1A5 / Production host:  Homo sapiens (human) / References: UniProt: Q00975 Homo sapiens (human) / References: UniProt: Q00975 |
|---|---|
| #2: Protein | Mass: 124692.469 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CACNA2D1, CACNL2A, CCHL2A, MHS3 / Production host: Homo sapiens (human) / Gene: CACNA2D1, CACNL2A, CCHL2A, MHS3 / Production host:  Homo sapiens (human) / References: UniProt: P54289 Homo sapiens (human) / References: UniProt: P54289 |
| #3: Protein | Mass: 65799.594 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CACNB1, CACNLB1 / Production host: Homo sapiens (human) / Gene: CACNB1, CACNLB1 / Production host:  Homo sapiens (human) / References: UniProt: Q02641 Homo sapiens (human) / References: UniProt: Q02641 |
-Sugars , 3 types, 11 molecules 
| #4: Polysaccharide | Source method: isolated from a genetically manipulated source #5: Polysaccharide | beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | Source method: isolated from a genetically manipulated source #11: Sugar | ChemComp-NAG / |
|---|
-Non-polymers , 5 types, 22 molecules 


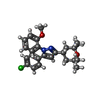





| #6: Chemical | ChemComp-R16 / #7: Chemical | #8: Chemical | ChemComp-PT5 / [( | #9: Chemical | ChemComp-6I7 / | #10: Chemical | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: CaV2.2-alpha2delta1-beta1 complex / Type: COMPLEX / Entity ID: #1-#3 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 9.6 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||
| CTF correction | Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 399567 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller















 PDBj
PDBj














