+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7sfr | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Unmethylated Mtb Ribosome 50S with SEQ-9 | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RIBOSOME/ANTIBIOTIC / unmethylated / Mtb ribosome / drug discovery / SEQ-9 / Structural Genomics / TB Structural Genomics Consortium / TBSGC / RIBOSOME-ANTIBIOTIC complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationlarge ribosomal subunit / ribosome biogenesis / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit ...large ribosomal subunit / ribosome biogenesis / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / zinc ion binding / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Xing, Z. / Cui, Z. / Zhang, J. / TB Structural Genomics Consortium (TBSGC) | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents. Authors: Jidong Zhang / Christine Lair / Christine Roubert / Kwame Amaning / María Belén Barrio / Yannick Benedetti / Zhicheng Cui / Zhongliang Xing / Xiaojun Li / Scott G Franzblau / Nicolas ...Authors: Jidong Zhang / Christine Lair / Christine Roubert / Kwame Amaning / María Belén Barrio / Yannick Benedetti / Zhicheng Cui / Zhongliang Xing / Xiaojun Li / Scott G Franzblau / Nicolas Baurin / Florence Bordon-Pallier / Cathy Cantalloube / Stephanie Sans / Sandra Silve / Isabelle Blanc / Laurent Fraisse / Alexey Rak / Lasse B Jenner / Gulnara Yusupova / Marat Yusupov / Junjie Zhang / Takushi Kaneko / T J Yang / Nader Fotouhi / Eric Nuermberger / Sandeep Tyagi / Fabrice Betoudji / Anna Upton / James C Sacchettini / Sophie Lagrange /   Abstract: The emergence of drug-resistant tuberculosis has created an urgent need for new anti-tubercular agents. Here, we report the discovery of a series of macrolides called sequanamycins with outstanding ...The emergence of drug-resistant tuberculosis has created an urgent need for new anti-tubercular agents. Here, we report the discovery of a series of macrolides called sequanamycins with outstanding in vitro and in vivo activity against Mycobacterium tuberculosis (Mtb). Sequanamycins are bacterial ribosome inhibitors that interact with the ribosome in a similar manner to classic macrolides like erythromycin and clarithromycin, but with binding characteristics that allow them to overcome the inherent macrolide resistance of Mtb. Structures of the ribosome with bound inhibitors were used to optimize sequanamycin to produce the advanced lead compound SEQ-9. SEQ-9 was efficacious in mouse models of acute and chronic TB as a single agent, and it demonstrated bactericidal activity in a murine TB infection model in combination with other TB drugs. These results support further investigation of this series as TB clinical candidates, with the potential for use in new regimens against drug-susceptible and drug-resistant TB. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7sfr.cif.gz 7sfr.cif.gz | 3.2 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7sfr.ent.gz pdb7sfr.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7sfr.json.gz 7sfr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sf/7sfr https://data.pdbj.org/pub/pdb/validation_reports/sf/7sfr ftp://data.pdbj.org/pub/pdb/validation_reports/sf/7sfr ftp://data.pdbj.org/pub/pdb/validation_reports/sf/7sfr | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  25100MC  7azoC  7azsC  7kgbC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
+50S ribosomal protein ... , 29 types, 29 molecules 012346CDEFGHJKLMNOPQRSTUVWXYZ
-RNA chain , 3 types, 3 molecules ABa
| #7: RNA chain | Mass: 1017862.688 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #8: RNA chain | Mass: 37097.238 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #32: RNA chain | Mass: 498415.750 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-30S ribosomal protein ... , 18 types, 18 molecules cdefghijklmnopqrst
| #33: Protein | Mass: 30071.018 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|---|
| #34: Protein | Mass: 23515.898 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #35: Protein | Mass: 22925.242 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #36: Protein | Mass: 10952.728 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #37: Protein | Mass: 17633.418 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #38: Protein | Mass: 14432.563 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #39: Protein | Mass: 16465.061 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #40: Protein | Mass: 11452.340 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #41: Protein | Mass: 14815.918 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #42: Protein | Mass: 13882.341 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #43: Protein | Mass: 14383.779 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #44: Protein | Mass: 6841.249 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #45: Protein | Mass: 10500.277 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #46: Protein | Mass: 17466.100 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #47: Protein | Mass: 14776.373 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #48: Protein | Mass: 9565.279 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #49: Protein | Mass: 10830.629 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
| #50: Protein | Mass: 9428.941 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
-Protein/peptide , 1 types, 1 molecules v
| #51: Protein/peptide | Mass: 2664.129 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  |
|---|
-Non-polymers , 3 types, 454 molecules 
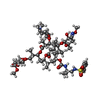



| #52: Chemical | ChemComp-ZN / #53: Chemical | ChemComp-WDP / | #54: Chemical | ChemComp-MG / |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Unmethylated Mth Ribosome with SEQ-9 / Type: RIBOSOME / Entity ID: #1-#51 / Source: NATURAL | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | |||||||||||||||||||||||||
| Source (natural) | Organism:  | |||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | |||||||||||||||||||||||||
| Buffer component |
| |||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES / Details: Monodispersed unmethylated Mtb ribosome with SEQ-9 | |||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R2/2 | |||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK III / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: OTHER |
| Electron lens | Mode: BRIGHT FIELD / Cs: 2.7 mm |
| Image recording | Electron dose: 42 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
| Image scans | Movie frames/image: 35 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||
| 3D reconstruction | Resolution: 2.6 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 719250 / Symmetry type: POINT | ||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL | ||||||||||||||||||||
| Atomic model building | PDB-ID: 7KGB Accession code: 7KGB / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller




 PDBj
PDBj
































