| Entry | Database: PDB / ID: 7qb3
|
|---|
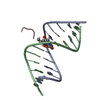
| Title | Solution structure of a lanthanide-binding DNA aptamer |
|---|
 Components Components | Lanthanide-binding aptamer |
|---|
 Keywords Keywords | DNA / aptamer / lanthanide |
|---|
| Function / homology | : / DNA / DNA (> 10) Function and homology information Function and homology information |
|---|
| Biological species | synthetic construct (others) |
|---|
| Method | SOLUTION NMR / simulated annealing |
|---|
 Authors Authors | Andralojc, W. / Gdaniec, Z. |
|---|
| Funding support |  Poland, 2items Poland, 2items | Organization | Grant number | Country |
|---|
| Polish National Science Centre | 2020/37/B/ST4/03182 |  Poland Poland | | Polish National Science Centre | 2018/31/D/ST4/01467 |  Poland Poland |
|
|---|
 Citation Citation |  Journal: Chemistry / Year: 2022 Journal: Chemistry / Year: 2022
Title: Solution Structure of a Lanthanide-binding DNA Aptamer Determined Using High Quality pseudocontact shift restraints.
Authors: Andralojc, W. / Wieruszewska, J. / Pasternak, K. / Gdaniec, Z. |
|---|
| History | | Deposition | Nov 18, 2021 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Dec 1, 2021 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Dec 21, 2022 | Group: Database references / Category: citation / citation_author
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.identifier_ORCID / _citation_author.name |
|---|
| Revision 1.2 | Jun 19, 2024 | Group: Data collection / Database references / Category: chem_comp_atom / chem_comp_bond / database_2 / Item: _database_2.pdbx_DOI |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Authors
Authors Poland, 2items
Poland, 2items  Citation
Citation Journal: Chemistry / Year: 2022
Journal: Chemistry / Year: 2022 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7qb3.cif.gz
7qb3.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7qb3.ent.gz
pdb7qb3.ent.gz PDB format
PDB format 7qb3.json.gz
7qb3.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 7qb3_validation.pdf.gz
7qb3_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 7qb3_full_validation.pdf.gz
7qb3_full_validation.pdf.gz 7qb3_validation.xml.gz
7qb3_validation.xml.gz 7qb3_validation.cif.gz
7qb3_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/qb/7qb3
https://data.pdbj.org/pub/pdb/validation_reports/qb/7qb3 ftp://data.pdbj.org/pub/pdb/validation_reports/qb/7qb3
ftp://data.pdbj.org/pub/pdb/validation_reports/qb/7qb3 Links
Links Assembly
Assembly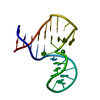
 Components
Components Sample preparation
Sample preparation Movie
Movie Controller
Controller









 PDBj
PDBj









































 HSQC
HSQC