[English] 日本語
 Yorodumi
Yorodumi- PDB-7opr: Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CB1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7opr | ||||||
|---|---|---|---|---|---|---|---|
| Title | Rab27a fusion with Slp2a-RBDa1 effector covalent adduct with CB1 in C123 | ||||||
 Components Components | Synaptotagmin-like protein 2,Ras-related protein Rab-27A | ||||||
 Keywords Keywords | EXOCYTOSIS / GTPase / Exosome / Acrylamide / Cysteine-reactive | ||||||
| Function / homology |  Function and homology information Function and homology informationmultivesicular body organization / cytotoxic T cell degranulation / positive regulation of constitutive secretory pathway / positive regulation of regulated secretory pathway / melanosome localization / natural killer cell degranulation / exosomal secretion / melanosome transport / Weibel-Palade body / neurexin family protein binding ...multivesicular body organization / cytotoxic T cell degranulation / positive regulation of constitutive secretory pathway / positive regulation of regulated secretory pathway / melanosome localization / natural killer cell degranulation / exosomal secretion / melanosome transport / Weibel-Palade body / neurexin family protein binding / exocytic vesicle / melanocyte differentiation / melanosome membrane / multivesicular body sorting pathway / RAB geranylgeranylation / myosin V binding / multivesicular body membrane / RAB GEFs exchange GTP for GDP on RABs / Regulation of MITF-M-dependent genes involved in pigmentation / complement-dependent cytotoxicity / vesicle docking involved in exocytosis / positive regulation of reactive oxygen species biosynthetic process / synaptic vesicle transport / Insulin processing / phosphatidylserine binding / exocytosis / positive regulation of exocytosis / antigen processing and presentation / protein secretion / photoreceptor outer segment / phosphatase binding / positive regulation of phagocytosis / vesicle-mediated transport / phosphatidylinositol-4,5-bisphosphate binding / small monomeric GTPase / G protein activity / secretory granule / intracellular protein transport / small GTPase binding / specific granule lumen / GDP binding / blood coagulation / melanosome / late endosome / lysosome / apical plasma membrane / protein domain specific binding / GTPase activity / dendrite / Neutrophil degranulation / positive regulation of gene expression / GTP binding / Golgi apparatus / extracellular exosome / extracellular region / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.32 Å MOLECULAR REPLACEMENT / Resolution: 2.32 Å | ||||||
 Authors Authors | Jamshidiha, M. / Tersa, M. / Lanyon-Hogg, T. / Perez-Dorado, I. / Sutherell, C.L. / De Vita, E. / Morgan, R.M.L. / Tate, E.W. / Cota, E. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Rsc Med Chem / Year: 2022 Journal: Rsc Med Chem / Year: 2022Title: Identification of the first structurally validated covalent ligands of the small GTPase RAB27A. Authors: Jamshidiha, M. / Lanyon-Hogg, T. / Sutherell, C.L. / Craven, G.B. / Tersa, M. / De Vita, E. / Brustur, D. / Perez-Dorado, I. / Hassan, S. / Petracca, R. / Morgan, R.M. / Sanz-Hernandez, M. / ...Authors: Jamshidiha, M. / Lanyon-Hogg, T. / Sutherell, C.L. / Craven, G.B. / Tersa, M. / De Vita, E. / Brustur, D. / Perez-Dorado, I. / Hassan, S. / Petracca, R. / Morgan, R.M. / Sanz-Hernandez, M. / Norman, J.C. / Armstrong, A. / Mann, D.J. / Cota, E. / Tate, E.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7opr.cif.gz 7opr.cif.gz | 113.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7opr.ent.gz pdb7opr.ent.gz | 85.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7opr.json.gz 7opr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7opr_validation.pdf.gz 7opr_validation.pdf.gz | 1.8 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7opr_full_validation.pdf.gz 7opr_full_validation.pdf.gz | 1.8 MB | Display | |
| Data in XML |  7opr_validation.xml.gz 7opr_validation.xml.gz | 22.9 KB | Display | |
| Data in CIF |  7opr_validation.cif.gz 7opr_validation.cif.gz | 31.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/op/7opr https://data.pdbj.org/pub/pdb/validation_reports/op/7opr ftp://data.pdbj.org/pub/pdb/validation_reports/op/7opr ftp://data.pdbj.org/pub/pdb/validation_reports/op/7opr | HTTPS FTP |
-Related structure data
| Related structure data |  7oppC  7opqC  3bc1S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 25857.162 Da / Num. of mol.: 2 / Mutation: Q78L,C188S Source method: isolated from a genetically manipulated source Details: The ligand CB1 is covalently bound to cysteine123. I have modelled the ligand CB1 already covalently bound to Cys123 into the peptide chain, for this reason there is a mismatch in the ...Details: The ligand CB1 is covalently bound to cysteine123. I have modelled the ligand CB1 already covalently bound to Cys123 into the peptide chain, for this reason there is a mismatch in the sequence. I am not sure how to fix this mismatch. I hope this information is clear.,The ligand CB1 is covalently bound to cysteine123. I have modelled the ligand CB1 already covalently bound to Cys123 into the peptide chain, for this reason there is a mismatch in the sequence. I am not sure how to fix this mismatch. I hope this information is clear. Source: (gene. exp.)  Homo sapiens (human) / Gene: SYTL2, KIAA1597, SGA72M, SLP2, SLP2A, RAB27A, RAB27 / Production host: Homo sapiens (human) / Gene: SYTL2, KIAA1597, SGA72M, SLP2, SLP2A, RAB27A, RAB27 / Production host:  References: UniProt: Q9HCH5, UniProt: P51159, small monomeric GTPase |
|---|
-Non-polymers , 5 types, 263 molecules 


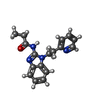





| #2: Chemical | | #3: Chemical | #4: Chemical | ChemComp-GOL / #5: Chemical |   Mass: 294.351 Da / Num. of mol.: 2 / Mutation: Q78L, C188S Mass: 294.351 Da / Num. of mol.: 2 / Mutation: Q78L, C188SSource method: isolated from a genetically manipulated source Formula: C17H18N4O Details: The ligand CB1 is covalently bound to cysteine123. I have modelled the ligand CB1 already covalently bound to Cys123 into the peptide chain, for this reason there is a mismatch in the ...Details: The ligand CB1 is covalently bound to cysteine123. I have modelled the ligand CB1 already covalently bound to Cys123 into the peptide chain, for this reason there is a mismatch in the sequence. I am not sure how to fix this mismatch. I hope this information is clear. Source: (gene. exp.)  Homo sapiens (human) / Gene: RAB27A, RAB27 / Production host: Homo sapiens (human) / Gene: RAB27A, RAB27 / Production host:  #6: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.69 Å3/Da / Density % sol: 54.28 % |
|---|---|
| Crystal grow | Temperature: 277.15 K / Method: vapor diffusion, sitting drop / pH: 6 Details: 0.15M Ammonium sulphate, 0.1M MES pH 6.0, 15% w/v PEG 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04 / Wavelength: 0.9795 Å / Beamline: I04 / Wavelength: 0.9795 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Jul 17, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 2.32→64.33 Å / Num. obs: 49537 / % possible obs: 99.82 % / Redundancy: 2 % / Biso Wilson estimate: 32.9 Å2 / CC1/2: 0.993 / CC star: 0.998 / Rmerge(I) obs: 0.05896 / Rpim(I) all: 0.05896 / Rrim(I) all: 0.08338 / Net I/σ(I): 9.77 |
| Reflection shell | Resolution: 2.32→2.403 Å / Redundancy: 2 % / Rmerge(I) obs: 0.3254 / Mean I/σ(I) obs: 2.23 / Num. unique obs: 4902 / CC1/2: 0.832 / CC star: 0.953 / Rpim(I) all: 0.3254 / Rrim(I) all: 0.4602 / % possible all: 99.67 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3BC1 Resolution: 2.32→64.33 Å / SU ML: 0.29 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 25.65 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 108.06 Å2 / Biso mean: 36.1718 Å2 / Biso min: 11.48 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.32→64.33 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0
|
 Movie
Movie Controller
Controller


 PDBj
PDBj













