| Entry | Database: PDB / ID: 7net
|
|---|
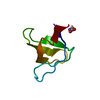
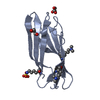
| Title | Crystal structure of the v-Src SH3 domain W95R-I96T mutant |
|---|
 Components Components | v-Src SH3 domain |
|---|
 Keywords Keywords | PROTEIN BINDING / beta barrel / SH3 domain |
|---|
| Function / homology | DI(HYDROXYETHYL)ETHER / TRIETHYLENE GLYCOL Function and homology information Function and homology information |
|---|
| Biological species |   Gallus gallus (chicken) Gallus gallus (chicken) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.5 Å molecular replacement / Resolution: 1.5 Å |
|---|
| Model details | Intertwined dimer |
|---|
 Authors Authors | Camara-Artigas, A. / Salinas-Garcia, M.C. |
|---|
| Funding support |  Spain, 1items Spain, 1items | Organization | Grant number | Country |
|---|
| Spanish Ministry of Economy and Competitiveness | BIO2016-78020-R |  Spain Spain |
|
|---|
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2021 Journal: Acta Crystallogr D Struct Biol / Year: 2021
Title: The impact of oncogenic mutations of the viral Src kinase on the structure and stability of the SH3 domain.
Authors: Salinas-Garcia, M.C. / Plaza-Garrido, M. / Camara-Artigas, A. |
|---|
| History | | Deposition | Feb 4, 2021 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Jun 2, 2021 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Jun 16, 2021 | Group: Database references / Category: citation / citation_author |
|---|
| Revision 1.2 | Jan 31, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| Revision 1.3 | Feb 11, 2026 | Group: Database references / Structure summary / Category: citation / citation_author / pdbx_entry_details / Item: _pdbx_entry_details.has_protein_modification |
|---|
|
|---|
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT /
MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.5 Å
molecular replacement / Resolution: 1.5 Å  Authors
Authors Spain, 1items
Spain, 1items  Citation
Citation Journal: Acta Crystallogr D Struct Biol / Year: 2021
Journal: Acta Crystallogr D Struct Biol / Year: 2021 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 7net.cif.gz
7net.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb7net.ent.gz
pdb7net.ent.gz PDB format
PDB format 7net.json.gz
7net.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ne/7net
https://data.pdbj.org/pub/pdb/validation_reports/ne/7net ftp://data.pdbj.org/pub/pdb/validation_reports/ne/7net
ftp://data.pdbj.org/pub/pdb/validation_reports/ne/7net

 Links
Links Assembly
Assembly
 Movie
Movie Controller
Controller












 PDBj
PDBj