[English] 日本語
 Yorodumi
Yorodumi- PDB-7lnf: 3'-deoxy modification at 3' end of RNA primer complex with guanos... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7lnf | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3'-deoxy modification at 3' end of RNA primer complex with guanosine dinucleotide ligand G(5')ppp(5')G | |||||||||||||||||||||||||||||||
 Components Components | RNA (5'-R(* Keywords KeywordsRNA / nonenzymatic RNA extension | Function / homology | : / 3'-deoxy-guanosine 5'-monophosphate / DIGUANOSINE-5'-TRIPHOSPHATE / Chem-LCC / Chem-LCG / Chem-LKC / RNA / RNA (> 10) |  Function and homology information Function and homology informationBiological species | synthetic construct (others) | Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.652 Å MOLECULAR REPLACEMENT / Resolution: 1.652 Å  Authors AuthorsFang, Z. / Giurgiu, C. / Szostak, J.W. | Funding support | |  United States, 2items United States, 2items
 Citation Citation Journal: Angew.Chem.Int.Ed.Engl. / Year: 2021 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2021Title: Structure-Activity Relationships in Nonenzymatic Template-Directed RNA Synthesis. Authors: Giurgiu, C. / Fang, Z. / Aitken, H.R.M. / Kim, S.C. / Pazienza, L. / Mittal, S. / Szostak, J.W. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7lnf.cif.gz 7lnf.cif.gz | 38.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7lnf.ent.gz pdb7lnf.ent.gz | 25.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7lnf.json.gz 7lnf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ln/7lnf https://data.pdbj.org/pub/pdb/validation_reports/ln/7lnf ftp://data.pdbj.org/pub/pdb/validation_reports/ln/7lnf ftp://data.pdbj.org/pub/pdb/validation_reports/ln/7lnf | HTTPS FTP |
|---|
-Related structure data
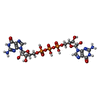
| Related structure data |  7kukC  7kulC  7kumC  7kunC  7kuoC 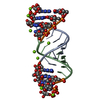 7kupC  7lneC  7lngC  5ueeS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||
| Unit cell |
| |||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-RNA chain , 1 types, 2 molecules AB
| #1: RNA chain | Mass: 4422.683 Da / Num. of mol.: 2 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|
-Non-polymers , 8 types, 145 molecules 


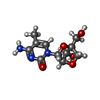










| #2: Chemical | ChemComp-LCC / [( #3: Chemical | #4: Chemical | #5: Chemical | #6: Chemical | #7: Chemical | ChemComp-MG / #8: Chemical | #9: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.98 Å3/Da / Density % sol: 58.67 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 6 Details: 0.02 M Magnesium sulfate hydrate, 0.002 M Cobalt (II) chloride hexahydrate, 0.05 M Sodium cacodylate trihydrate pH 6.0, 25% v/v (+/-)-2-Methyl-2,4-pentanediol, 0.0005 M Spermine |
-Data collection
| Diffraction | Mean temperature: 99 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.2.2 / Wavelength: 1 Å / Beamline: 8.2.2 / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Dec 11, 2020 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.65→50 Å / Num. obs: 13195 / % possible obs: 99.8 % / Redundancy: 10 % / CC1/2: 1 / CC star: 1 / Rmerge(I) obs: 0.084 / Rpim(I) all: 0.03 / Rrim(I) all: 0.09 / Χ2: 0.935 / Net I/σ(I): 23.1 |
| Reflection shell | Resolution: 1.65→1.68 Å / Redundancy: 8 % / Rmerge(I) obs: 0.393 / Mean I/σ(I) obs: 4.5 / Num. unique obs: 658 / CC1/2: 0.987 / CC star: 0.997 / Rpim(I) all: 0.142 / Rrim(I) all: 0.418 / Χ2: 0.732 / % possible all: 99.4 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5UEE Resolution: 1.652→27.748 Å / Cor.coef. Fo:Fc: 0.943 / Cor.coef. Fo:Fc free: 0.925 / SU B: 1.925 / SU ML: 0.066 / Cross valid method: THROUGHOUT / ESU R: 0.118 / ESU R Free: 0.114 Details: Hydrogens have been added in their riding positions
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 15.582 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.652→27.748 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj





































