[English] 日本語
 Yorodumi
Yorodumi- PDB-7kyv: Crystal structure of Plasmodium falciparum dihydroorotate dehydro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7kyv | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM634 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(4-(trifluoromethyl)benzyl)-1H-pyrrole-2-carboxamide) | ||||||
 Components Components | Dihydroorotate dehydrogenase (quinone), mitochondrial | ||||||
 Keywords Keywords | OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR / Alpha-Beta Barrel / OXIDOREDUCTASE / OXIDOREDUCTASE-OXIDOREDUCTASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationPyrimidine biosynthesis / pyrimidine ribonucleotide biosynthetic process / dihydroorotate dehydrogenase (quinone) / dihydroorotate dehydrogenase (quinone) activity / dihydroorotate dehydrogenase activity / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process / mitochondrial inner membrane Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.4 Å MOLECULAR REPLACEMENT / Resolution: 2.4 Å | ||||||
 Authors Authors | Deng, X. / Phillips, M. / Tomchick, D. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2021 Journal: J.Med.Chem. / Year: 2021Title: Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series. Authors: Palmer, M.J. / Deng, X. / Watts, S. / Krilov, G. / Gerasyuto, A. / Kokkonda, S. / El Mazouni, F. / White, J. / White, K.L. / Striepen, J. / Bath, J. / Schindler, K.A. / Yeo, T. / ...Authors: Palmer, M.J. / Deng, X. / Watts, S. / Krilov, G. / Gerasyuto, A. / Kokkonda, S. / El Mazouni, F. / White, J. / White, K.L. / Striepen, J. / Bath, J. / Schindler, K.A. / Yeo, T. / Shackleford, D.M. / Mok, S. / Deni, I. / Lawong, A. / Huang, A. / Chen, G. / Wang, W. / Jayaseelan, J. / Katneni, K. / Patil, R. / Saunders, J. / Shahi, S.P. / Chittimalla, R. / Angulo-Barturen, I. / Jimenez-Diaz, M.B. / Wittlin, S. / Tumwebaze, P.K. / Rosenthal, P.J. / Cooper, R.A. / Aguiar, A.C.C. / Guido, R.V.C. / Pereira, D.B. / Mittal, N. / Winzeler, E.A. / Tomchick, D.R. / Laleu, B. / Burrows, J.N. / Rathod, P.K. / Fidock, D.A. / Charman, S.A. / Phillips, M.A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7kyv.cif.gz 7kyv.cif.gz | 281.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7kyv.ent.gz pdb7kyv.ent.gz | 191.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7kyv.json.gz 7kyv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ky/7kyv https://data.pdbj.org/pub/pdb/validation_reports/ky/7kyv ftp://data.pdbj.org/pub/pdb/validation_reports/ky/7kyv ftp://data.pdbj.org/pub/pdb/validation_reports/ky/7kyv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7kykC  7kyyC  7kz4C  7kzyC  7l01C  7l0kC  3i65S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 43162.539 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Strain: isolate 3D7 / Gene: PFF0160c / Production host:  References: UniProt: Q08210, dihydroorotate dehydrogenase (quinone) |
|---|
-Non-polymers , 5 types, 49 molecules 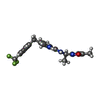







| #2: Chemical | ChemComp-XBY / | ||
|---|---|---|---|
| #3: Chemical | ChemComp-FMN / | ||
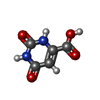
| #4: Chemical | ChemComp-ORO / | ||
| #5: Chemical | ChemComp-GOL / #6: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.15 Å3/Da / Density % sol: 61 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 5.1 Details: 0.16 M AmSO4, 18% v/v PEG4000, 100 mM Sodium acetate pH 5.1, 24% v/v Glycerol |
-Data collection
| Diffraction | Mean temperature: 100 K / Ambient temp details: liquid N2 / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.97943 Å / Beamline: 19-ID / Wavelength: 0.97943 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Jun 15, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97943 Å / Relative weight: 1 |
| Reflection | Resolution: 2.4→42.64 Å / Num. obs: 22433 / % possible obs: 100 % / Redundancy: 9.6 % / Biso Wilson estimate: 32.6 Å2 / Rpim(I) all: 0.017 / Net I/σ(I): 46.22 |
| Reflection shell | Resolution: 2.4→2.44 Å / Mean I/σ(I) obs: 1.1 / Num. unique obs: 1121 / Rpim(I) all: 0.721 / % possible all: 100 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3I65 Resolution: 2.4→42.64 Å / Cross valid method: FREE R-VALUE / σ(F): 16.82 / Phase error: 22.7417 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 46.6 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4→42.64 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.4→42.64 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj



