[English] 日本語
 Yorodumi
Yorodumi- PDB-7kpb: Human TNF-alpha TNFR1 complex bound to conformationally selective... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7kpb | ||||||
|---|---|---|---|---|---|---|---|
| Title | Human TNF-alpha TNFR1 complex bound to conformationally selective antibody | ||||||
 Components Components |
| ||||||
 Keywords Keywords | CYTOKINE/Immune System / Tumour necrosis factor alpha / TNF / asymmetric / protein-protein inhibitor / CYTOKINE / Fragment antibody / Fab / CYTOKINE-Immune System complex | ||||||
| Function / homology |  Function and homology information Function and homology information: / tumor necrosis factor receptor superfamily complex / pulmonary valve development / negative regulation of L-glutamate import across plasma membrane / negative regulation of bile acid secretion / response to Gram-negative bacterium / positive regulation of neutrophil activation / positive regulation of fractalkine production / negative regulation of branching involved in lung morphogenesis / positive regulation of interleukin-33 production ...: / tumor necrosis factor receptor superfamily complex / pulmonary valve development / negative regulation of L-glutamate import across plasma membrane / negative regulation of bile acid secretion / response to Gram-negative bacterium / positive regulation of neutrophil activation / positive regulation of fractalkine production / negative regulation of branching involved in lung morphogenesis / positive regulation of interleukin-33 production / positive regulation of lipid metabolic process / positive regulation of blood microparticle formation / aortic valve development / positive regulation of chronic inflammatory response to antigenic stimulus / tumor necrosis factor receptor activity / negative regulation of extracellular matrix constituent secretion / response to 3,3',5-triiodo-L-thyronine / positive regulation of protein transport / positive regulation of leukocyte adhesion to arterial endothelial cell / positive regulation of vitamin D biosynthetic process / positive regulation of apoptotic process involved in morphogenesis / positive regulation of translational initiation by iron / : / regulation of endothelial cell apoptotic process / chronic inflammatory response to antigenic stimulus / response to macrophage colony-stimulating factor / regulation of branching involved in salivary gland morphogenesis / negative regulation of protein-containing complex disassembly / tumor necrosis factor binding / positive regulation of humoral immune response mediated by circulating immunoglobulin / negative regulation of myelination / positive regulation of hair follicle development / response to gold nanoparticle / TNFs bind their physiological receptors / TNF signaling / negative regulation of vascular wound healing / negative regulation of cytokine production involved in immune response / negative regulation of amyloid-beta clearance / positive regulation of interleukin-18 production / inflammatory response to wounding / negative regulation of cardiac muscle hypertrophy / death receptor agonist activity / positive regulation of action potential / epithelial cell proliferation involved in salivary gland morphogenesis / toll-like receptor 3 signaling pathway / negative regulation of D-glucose import across plasma membrane / vascular endothelial growth factor production / embryonic digestive tract development / positive regulation of fever generation / leukocyte migration involved in inflammatory response / response to fructose / positive regulation of neuroinflammatory response / positive regulation of synoviocyte proliferation / positive regulation of calcineurin-NFAT signaling cascade / necroptotic signaling pathway / positive regulation of mononuclear cell migration / leukocyte tethering or rolling / positive regulation of hepatocyte proliferation / negative regulation of myoblast differentiation / positive regulation of protein-containing complex disassembly / regulation of establishment of endothelial barrier / endothelial cell apoptotic process / negative regulation of oxidative phosphorylation / macrophage activation involved in immune response / cellular response to toxic substance / regulation of tumor necrosis factor-mediated signaling pathway / positive regulation of protein localization to cell surface / positive regulation of osteoclast differentiation / tumor necrosis factor receptor binding / negative regulation of systemic arterial blood pressure / positive regulation of heterotypic cell-cell adhesion / positive regulation of macrophage derived foam cell differentiation / positive regulation of cytokine production involved in inflammatory response / regulation of immunoglobulin production / negative regulation of mitotic cell cycle / positive regulation of programmed cell death / positive regulation of podosome assembly / positive regulation of chemokine (C-X-C motif) ligand 2 production / positive regulation of membrane protein ectodomain proteolysis / regulation of fat cell differentiation / response to L-glutamate / regulation of canonical NF-kappaB signal transduction / positive regulation of leukocyte adhesion to vascular endothelial cell / positive regulation of extrinsic apoptotic signaling pathway / TNFR1-mediated ceramide production / TNFR1-induced proapoptotic signaling / regulation of metabolic process / regulation of reactive oxygen species metabolic process / positive regulation of DNA biosynthetic process / prostaglandin metabolic process / negative regulation of heart rate / positive regulation of amyloid-beta formation / negative regulation of bicellular tight junction assembly / negative regulation of viral genome replication / response to isolation stress / negative regulation of fat cell differentiation / regulation of synapse organization / negative regulation of endothelial cell proliferation / positive regulation of JUN kinase activity / positive regulation of MAP kinase activity Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3 Å MOLECULAR REPLACEMENT / Resolution: 3 Å | ||||||
 Authors Authors | Fox III, D. / Conrady, D.G. / Lowe, M. / Ceska, T. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: A conformation-selective monoclonal antibody against a small molecule-stabilised signalling-deficient form of TNF Authors: Lightwood, D.J. / Munro, R.J. / Porter, J. / McMillan, D. / Carrington, B. / Turner, A. / Scott-Tucker, A. / Hickford, E.S. / Schmidt, A. / Fox III, D. / Maloney, A. / Ceska, T. / Bourne, T. ...Authors: Lightwood, D.J. / Munro, R.J. / Porter, J. / McMillan, D. / Carrington, B. / Turner, A. / Scott-Tucker, A. / Hickford, E.S. / Schmidt, A. / Fox III, D. / Maloney, A. / Ceska, T. / Bourne, T. / O'Connell, J. / Lawson, A.D.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7kpb.cif.gz 7kpb.cif.gz | 233.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7kpb.ent.gz pdb7kpb.ent.gz | 180.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7kpb.json.gz 7kpb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7kpb_validation.pdf.gz 7kpb_validation.pdf.gz | 832.9 KB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7kpb_full_validation.pdf.gz 7kpb_full_validation.pdf.gz | 840.9 KB | Display | |
| Data in XML |  7kpb_validation.xml.gz 7kpb_validation.xml.gz | 40.3 KB | Display | |
| Data in CIF |  7kpb_validation.cif.gz 7kpb_validation.cif.gz | 54.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kp/7kpb https://data.pdbj.org/pub/pdb/validation_reports/kp/7kpb ftp://data.pdbj.org/pub/pdb/validation_reports/kp/7kpb ftp://data.pdbj.org/pub/pdb/validation_reports/kp/7kpb | HTTPS FTP |
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Tumor necrosis ... , 2 types, 5 molecules ABCFE
| #1: Protein | Mass: 17457.736 Da / Num. of mol.: 3 / Mutation: N25D, C153S Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TNF, TNFA, TNFSF2 / Plasmid: pBAD_Smt3_Bam / Production host: Homo sapiens (human) / Gene: TNF, TNFA, TNFSF2 / Plasmid: pBAD_Smt3_Bam / Production host:  #2: Protein | Mass: 16196.158 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: TNFRSF1A, TNFAR, TNFR1 / Plasmid: pEMB50 / Production host: Homo sapiens (human) / Gene: TNFRSF1A, TNFAR, TNFR1 / Plasmid: pEMB50 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P19438 Trichoplusia ni (cabbage looper) / References: UniProt: P19438 |
|---|
-Antibody , 2 types, 2 molecules LH
| #3: Antibody | Mass: 23649.170 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human) |
|---|---|
| #4: Antibody | Mass: 24066.783 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) Homo sapiens (human) |
-Sugars , 1 types, 1 molecules 
| #7: Sugar | ChemComp-NAG / |
|---|
-Non-polymers , 3 types, 30 molecules 
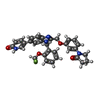



| #5: Chemical | ChemComp-GOL / #6: Chemical | ChemComp-D84 / | #8: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.91 Å3/Da / Density % sol: 57.72 % |
|---|---|
| Crystal grow | Temperature: 289 K / Method: vapor diffusion, sitting drop / pH: 7 Details: 0.1M HEPES, pH7.0, 10% w/v PEG6,000 and cryo-protected in glycerol performed in 5-10-15% steps. Vapor diffusion, sitting drop, temperature 289K PH range: 7 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 21-ID-F / Wavelength: 0.9787 Å / Beamline: 21-ID-F / Wavelength: 0.9787 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Jun 8, 2016 / Details: Beryllium Lenses | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Diamond [111] / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.9787 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 3→50 Å / Num. obs: 32445 / % possible obs: 99.9 % / Observed criterion σ(I): -3 / Redundancy: 12.1 % / Biso Wilson estimate: 71.095 Å2 / CC1/2: 0.996 / Rmerge(I) obs: 0.163 / Rrim(I) all: 0.171 / Χ2: 0.958 / Net I/σ(I): 13.95 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: unpublished structure Resolution: 3→49.132 Å / SU ML: 0.41 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 25.73 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 175.92 Å2 / Biso mean: 69.5983 Å2 / Biso min: 23.1 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3→49.132 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj













