[English] 日本語
 Yorodumi
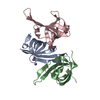
Yorodumi- PDB-7e5e: Crystal structure of GDP-bound GNAS in complex with the cyclic pe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7e5e | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of GDP-bound GNAS in complex with the cyclic peptide inhibitor GD20 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / G protein / GNAS / adenylyl cyclase / inactive state / inhibitor / mRNA display / RaPID / cyclic peptide / GD20 | ||||||
| Function / homology |  Function and homology information Function and homology informationPKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway ...PKA activation in glucagon signalling / developmental growth / hair follicle placode formation / D1 dopamine receptor binding / intracellular transport / vascular endothelial cell response to laminar fluid shear stress / renal water homeostasis / activation of adenylate cyclase activity / Hedgehog 'off' state / adenylate cyclase-activating adrenergic receptor signaling pathway / regulation of insulin secretion / cellular response to glucagon stimulus / adenylate cyclase activator activity / trans-Golgi network membrane / negative regulation of inflammatory response to antigenic stimulus / bone development / platelet aggregation / cognition / G-protein beta/gamma-subunit complex binding / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / sensory perception of smell / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / positive regulation of cold-induced thermogenesis / G protein activity / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / G protein-coupled receptor signaling pathway / GTPase activity / GTP binding / extracellular exosome / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.95 Å MOLECULAR REPLACEMENT / Resolution: 1.95 Å | ||||||
 Authors Authors | Hu, Q. / Dai, S. / Shokat, K.M. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: State-selective modulation of heterotrimeric G alpha s signaling with macrocyclic peptides. Authors: Dai, S.A. / Hu, Q. / Gao, R. / Blythe, E.E. / Touhara, K.K. / Peacock, H. / Zhang, Z. / von Zastrow, M. / Suga, H. / Shokat, K.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7e5e.cif.gz 7e5e.cif.gz | 346.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7e5e.ent.gz pdb7e5e.ent.gz | 238.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7e5e.json.gz 7e5e.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7e5e_validation.pdf.gz 7e5e_validation.pdf.gz | 3.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7e5e_full_validation.pdf.gz 7e5e_full_validation.pdf.gz | 3.3 MB | Display | |
| Data in XML |  7e5e_validation.xml.gz 7e5e_validation.xml.gz | 52.6 KB | Display | |
| Data in CIF |  7e5e_validation.cif.gz 7e5e_validation.cif.gz | 71.7 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/e5/7e5e https://data.pdbj.org/pub/pdb/validation_reports/e5/7e5e ftp://data.pdbj.org/pub/pdb/validation_reports/e5/7e5e ftp://data.pdbj.org/pub/pdb/validation_reports/e5/7e5e | HTTPS FTP |
-Related structure data
| Related structure data |  7bphC  6au6S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| 3 | 
| ||||||||||||
| 4 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 40512.871 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GNAS, GNAS1, GSP / Production host: Homo sapiens (human) / Gene: GNAS, GNAS1, GSP / Production host:  #2: Protein/peptide | Mass: 1854.182 Da / Num. of mol.: 4 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) #3: Chemical | ChemComp-GDP / #4: Chemical | ChemComp-CL / #5: Water | ChemComp-HOH / | Has ligand of interest | Y | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.18 Å3/Da / Density % sol: 43.62 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 8.2 / Details: 0.1M Tris 8.2, 26% PEG 4000, 0.8M LiCl |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 8.2.1 / Wavelength: 0.999965 Å / Beamline: 8.2.1 / Wavelength: 0.999965 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Dec 14, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.999965 Å / Relative weight: 1 |
| Reflection | Resolution: 1.95→50 Å / Num. obs: 95974 / % possible obs: 97 % / Redundancy: 3.4 % / Biso Wilson estimate: 19.71 Å2 / CC1/2: 0.991 / Rmerge(I) obs: 0.081 / Rpim(I) all: 0.059 / Net I/σ(I): 11.4 |
| Reflection shell | Resolution: 1.95→1.98 Å / Rmerge(I) obs: 1.202 / Mean I/σ(I) obs: 0.815 / Num. unique obs: 4489 / CC1/2: 0.422 / Rpim(I) all: 0.762 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6AU6 Resolution: 1.95→48.54 Å / SU ML: 0.2546 / Cross valid method: FREE R-VALUE / σ(F): 1.96 / Phase error: 29.8601 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 29.01 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.95→48.54 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj















