[English] 日本語
 Yorodumi
Yorodumi- PDB-7a8p: Structure of human mitochondrial RNA polymerase in complex with I... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7a8p | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | ||||||||||||||||||||||||||||||||||||||||||||||||
 Components Components | DNA-directed RNA polymerase, mitochondrial | ||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | TRANSCRIPTION / Mitochondria / Polymerase / Inhibitor | ||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / mitochondrial transcription / mitochondrial nucleoid / Transcriptional activation of mitochondrial biogenesis / DNA-directed RNA polymerase ...Mitochondrial transcription initiation / mitochondrial DNA-directed RNA polymerase complex / mitochondrial promoter sequence-specific DNA binding / transcription initiation at mitochondrial promoter / Strand-asynchronous mitochondrial DNA replication / mitochondrial DNA replication / mitochondrial transcription / mitochondrial nucleoid / Transcriptional activation of mitochondrial biogenesis / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / 3'-5'-RNA exonuclease activity / sequence-specific DNA binding / mitochondrial matrix / protein-containing complex / mitochondrion / RNA binding Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Hillen, H.S. / Bonekamp, N. / Peter, B. / Felser, A. / Bergbrede, T. / Choidas, A. / Horn, M. / Unger, A. / di Lucrezia, R. / Atanassov, I. ...Hillen, H.S. / Bonekamp, N. / Peter, B. / Felser, A. / Bergbrede, T. / Choidas, A. / Horn, M. / Unger, A. / di Lucrezia, R. / Atanassov, I. / Li, X. / Koch, U. / Menninger, S. / Boros, J. / Habenberger, P. / Giavalisco, P. / Cramer, P. / Denzel, M. / Nussbaumer, P. / Klebl, B. / Falkenberg, M. / Gustafsson, C.M. / Larsson, N.G. | ||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  Sweden, Sweden,  Germany, 15items Germany, 15items
| ||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Small-molecule inhibitors of human mitochondrial DNA transcription. Authors: Nina A Bonekamp / Bradley Peter / Hauke S Hillen / Andrea Felser / Tim Bergbrede / Axel Choidas / Moritz Horn / Anke Unger / Raffaella Di Lucrezia / Ilian Atanassov / Xinping Li / Uwe Koch / ...Authors: Nina A Bonekamp / Bradley Peter / Hauke S Hillen / Andrea Felser / Tim Bergbrede / Axel Choidas / Moritz Horn / Anke Unger / Raffaella Di Lucrezia / Ilian Atanassov / Xinping Li / Uwe Koch / Sascha Menninger / Joanna Boros / Peter Habenberger / Patrick Giavalisco / Patrick Cramer / Martin S Denzel / Peter Nussbaumer / Bert Klebl / Maria Falkenberg / Claes M Gustafsson / Nils-Göran Larsson /    Abstract: Altered expression of mitochondrial DNA (mtDNA) occurs in ageing and a range of human pathologies (for example, inborn errors of metabolism, neurodegeneration and cancer). Here we describe first-in- ...Altered expression of mitochondrial DNA (mtDNA) occurs in ageing and a range of human pathologies (for example, inborn errors of metabolism, neurodegeneration and cancer). Here we describe first-in-class specific inhibitors of mitochondrial transcription (IMTs) that target the human mitochondrial RNA polymerase (POLRMT), which is essential for biogenesis of the oxidative phosphorylation (OXPHOS) system. The IMTs efficiently impair mtDNA transcription in a reconstituted recombinant system and cause a dose-dependent inhibition of mtDNA expression and OXPHOS in cell lines. To verify the cellular target, we performed exome sequencing of mutagenized cells and identified a cluster of amino acid substitutions in POLRMT that cause resistance to IMTs. We obtained a cryo-electron microscopy (cryo-EM) structure of POLRMT bound to an IMT, which further defined the allosteric binding site near the active centre cleft of POLRMT. The growth of cancer cells and the persistence of therapy-resistant cancer stem cells has previously been reported to depend on OXPHOS, and we therefore investigated whether IMTs have anti-tumour effects. Four weeks of oral treatment with an IMT is well-tolerated in mice and does not cause OXPHOS dysfunction or toxicity in normal tissues, despite inducing a strong anti-tumour response in xenografts of human cancer cells. In summary, IMTs provide a potent and specific chemical biology tool to study the role of mtDNA expression in physiology and disease. | ||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7a8p.cif.gz 7a8p.cif.gz | 181.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7a8p.ent.gz pdb7a8p.ent.gz | 135 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7a8p.json.gz 7a8p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7a8p_validation.pdf.gz 7a8p_validation.pdf.gz | 1.2 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7a8p_full_validation.pdf.gz 7a8p_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  7a8p_validation.xml.gz 7a8p_validation.xml.gz | 36.7 KB | Display | |
| Data in CIF |  7a8p_validation.cif.gz 7a8p_validation.cif.gz | 55.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/a8/7a8p https://data.pdbj.org/pub/pdb/validation_reports/a8/7a8p ftp://data.pdbj.org/pub/pdb/validation_reports/a8/7a8p ftp://data.pdbj.org/pub/pdb/validation_reports/a8/7a8p | HTTPS FTP |
-Related structure data
| Related structure data |  11679MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 129803.094 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: POLRMT / Plasmid: ProEx-His6TEVD104mtRNAP / Production host: Homo sapiens (human) / Gene: POLRMT / Plasmid: ProEx-His6TEVD104mtRNAP / Production host:  |
|---|---|
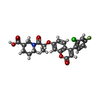
| #2: Chemical | ChemComp-R4Q / ( |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Human POLRMT (lacking residues 1-104) in complex with IMT inhibitor. Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.128 MDa / Experimental value: NO | ||||||||||||||||||||
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Mitochondria Homo sapiens (human) / Organelle: Mitochondria | ||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||
| Buffer solution | pH: 8 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||
| Specimen support | Grid material: GOLD / Grid type: UltrAuFoil R2/2 | ||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS / Details: Stage was tilted 40 deg. |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 105000 X / Nominal defocus max: 3000 nm / Nominal defocus min: 500 nm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 36.25 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
| EM imaging optics | Energyfilter name: GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1446981 | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 193651 / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: OTHER / Space: REAL | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 4BOC Pdb chain-ID: A / Accession code: 4BOC / Source name: PDB / Type: experimental model |
 Movie
Movie Controller
Controller












 PDBj
PDBj