+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 6zg0 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
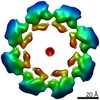
| タイトル | SARM1 SAM1-2 domains | |||||||||||||||||||||||||||
 要素 要素 | NAD(+) hydrolase SARM1 | |||||||||||||||||||||||||||
 キーワード キーワード | HYDROLASE / NADase / SAM domain | |||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報extrinsic component of synaptic membrane / negative regulation of MyD88-independent toll-like receptor signaling pathway / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase ...extrinsic component of synaptic membrane / negative regulation of MyD88-independent toll-like receptor signaling pathway / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / protein localization to mitochondrion / NAD+ nucleosidase activity, cyclic ADP-ribose generating / nervous system process / 加水分解酵素; 糖加水分解酵素; N-グリコシル化合物加水分解酵素 / regulation of dendrite morphogenesis / response to glucose / response to axon injury / regulation of neuron apoptotic process / signaling adaptor activity / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / IKK complex recruitment mediated by RIP1 / neuromuscular junction / nervous system development / microtubule / mitochondrial outer membrane / cell differentiation / axon / innate immune response / dendrite / synapse / glutamatergic synapse / cell surface / signal transduction / protein-containing complex / mitochondrion / identical protein binding / cytoplasm / cytosol 類似検索 - 分子機能 | |||||||||||||||||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||||||||||||||||||||
| 手法 | 電子顕微鏡法 / 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 7.7 Å | |||||||||||||||||||||||||||
 データ登録者 データ登録者 | Sporny, M. / Guez-Haddad, J. / Khazma, T. / Yaron, A. / Dessau, M. / Mim, C. / Isupov, M.N. / Zalk, R. / Hons, M. / Opatowsky, Y. | |||||||||||||||||||||||||||
| 資金援助 |  イスラエル, 2件 イスラエル, 2件
| |||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Elife / 年: 2020 ジャーナル: Elife / 年: 2020タイトル: Structural basis for SARM1 inhibition and activation under energetic stress. 著者: Michael Sporny / Julia Guez-Haddad / Tami Khazma / Avraham Yaron / Moshe Dessau / Yoel Shkolnisky / Carsten Mim / Michail N Isupov / Ran Zalk / Michael Hons / Yarden Opatowsky /     要旨: SARM1, an executor of axonal degeneration, displays NADase activity that depletes the key cellular metabolite, NAD+, in response to nerve injury. The basis of SARM1 inhibition and its activation ...SARM1, an executor of axonal degeneration, displays NADase activity that depletes the key cellular metabolite, NAD+, in response to nerve injury. The basis of SARM1 inhibition and its activation under stress conditions are still unknown. Here, we present cryo-EM maps of SARM1 at 2.9 and 2.7 Å resolutions. These indicate that SARM1 homo-octamer avoids premature activation by assuming a packed conformation, with ordered inner and peripheral rings, that prevents dimerization and activation of the catalytic domains. This inactive conformation is stabilized by binding of SARM1's own substrate NAD+ in an allosteric location, away from the catalytic sites. This model was validated by mutagenesis of the allosteric site, which led to constitutively active SARM1. We propose that the reduction of cellular NAD+ concentration contributes to the disassembly of SARM1's peripheral ring, which allows formation of active NADase domain dimers, thereby further depleting NAD+ to cause an energetic catastrophe and cell death. | |||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  6zg0.cif.gz 6zg0.cif.gz | 310.4 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb6zg0.ent.gz pdb6zg0.ent.gz | 222 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  6zg0.json.gz 6zg0.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  6zg0_validation.pdf.gz 6zg0_validation.pdf.gz | 971.1 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  6zg0_full_validation.pdf.gz 6zg0_full_validation.pdf.gz | 985.9 KB | 表示 | |
| XML形式データ |  6zg0_validation.xml.gz 6zg0_validation.xml.gz | 39.7 KB | 表示 | |
| CIF形式データ |  6zg0_validation.cif.gz 6zg0_validation.cif.gz | 57.2 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/zg/6zg0 https://data.pdbj.org/pub/pdb/validation_reports/zg/6zg0 ftp://data.pdbj.org/pub/pdb/validation_reports/zg/6zg0 ftp://data.pdbj.org/pub/pdb/validation_reports/zg/6zg0 | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
|
|---|---|
| 1 |
|
- 要素
要素
| #1: タンパク質 | 分子量: 79971.258 Da / 分子数: 8 / 由来タイプ: 組換発現 / 由来: (組換発現)  Homo sapiens (ヒト) / 遺伝子: SARM1, KIAA0524, SAMD2, SARM / 細胞株 (発現宿主): HEK293F / 発現宿主: Homo sapiens (ヒト) / 遺伝子: SARM1, KIAA0524, SAMD2, SARM / 細胞株 (発現宿主): HEK293F / 発現宿主:  Homo sapiens (ヒト) Homo sapiens (ヒト)参照: UniProt: Q6SZW1, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase, 加水分解酵素; 糖加水分解酵素; N-グリコシル化合物加水分解酵素 #2: 化合物 | ChemComp-EDO / #3: 化合物 | ChemComp-BME / #4: 化合物 | #5: 化合物 | ChemComp-PGE / | 研究の焦点であるリガンドがあるか | N | Has protein modification | N | |
|---|
-実験情報
-実験
| 実験 | 手法: 電子顕微鏡法 |
|---|---|
| EM実験 | 試料の集合状態: PARTICLE / 3次元再構成法: 単粒子再構成法 |
- 試料調製
試料調製
| 構成要素 | 名称: hSARM1 / タイプ: COMPLEX / Entity ID: #1 / 由来: RECOMBINANT |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 由来(組換発現) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 緩衝液 | pH: 8.8 |
| 試料 | 包埋: NO / シャドウイング: NO / 染色: NO / 凍結: YES |
| 急速凍結 | 凍結剤: ETHANE |
- 電子顕微鏡撮影
電子顕微鏡撮影
| 実験機器 |  モデル: Tecnai Polara / 画像提供: FEI Company |
|---|---|
| 顕微鏡 | モデル: FEI POLARA 300 |
| 電子銃 | 電子線源:  FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM |
| 電子レンズ | モード: BRIGHT FIELD |
| 撮影 | 電子線照射量: 80 e/Å2 フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) |
- 解析
解析
| CTF補正 | タイプ: PHASE FLIPPING AND AMPLITUDE CORRECTION |
|---|---|
| 3次元再構成 | 解像度: 7.7 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 粒子像の数: 5410 / 対称性のタイプ: POINT |
 ムービー
ムービー コントローラー
コントローラー














 PDBj
PDBj













