[English] 日本語
 Yorodumi
Yorodumi- PDB-6ws5: Rational drug design of phenazopyridine derivatives as novel inhi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ws5 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Rational drug design of phenazopyridine derivatives as novel inhibitors of Rev1-CT | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN BINDING/Transferase / scaffold / complex / translesion synethesis / DNA damage response / DNA damage tolerance / PROTEIN BINDING / PROTEIN BINDING-Transferase complex | ||||||
| Function / homology |  Function and homology information Function and homology informationsomatic diversification of immunoglobulins involved in immune response / DNA damage response, signal transduction resulting in transcription / deoxycytidyl transferase activity / zeta DNA polymerase complex / positive regulation of isotype switching / negative regulation of transcription by competitive promoter binding / negative regulation of cell-cell adhesion mediated by cadherin / JUN kinase binding / negative regulation of epithelial to mesenchymal transition / error-free translesion synthesis ...somatic diversification of immunoglobulins involved in immune response / DNA damage response, signal transduction resulting in transcription / deoxycytidyl transferase activity / zeta DNA polymerase complex / positive regulation of isotype switching / negative regulation of transcription by competitive promoter binding / negative regulation of cell-cell adhesion mediated by cadherin / JUN kinase binding / negative regulation of epithelial to mesenchymal transition / error-free translesion synthesis / positive regulation of double-strand break repair via nonhomologous end joining / mitotic spindle assembly checkpoint signaling / negative regulation of ubiquitin protein ligase activity / telomere maintenance in response to DNA damage / positive regulation of peptidyl-serine phosphorylation / error-prone translesion synthesis / negative regulation of double-strand break repair via homologous recombination / response to UV / actin filament organization / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / regulation of cell growth / Termination of translesion DNA synthesis / negative regulation of canonical Wnt signaling pathway / double-strand break repair via homologous recombination / negative regulation of protein catabolic process / DNA-templated DNA replication / spindle / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / transcription corepressor activity / double-strand break repair / site of double-strand break / chromosome / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / damaged DNA binding / RNA polymerase II-specific DNA-binding transcription factor binding / DNA-directed DNA polymerase activity / DNA replication / cell division / nucleotide binding / positive regulation of DNA-templated transcription / chromatin / nucleolus / negative regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / nucleoplasm / metal ion binding / nucleus / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.472 Å MOLECULAR REPLACEMENT / Resolution: 2.472 Å | ||||||
 Authors Authors | McPherson, K.S. / Korzhnev, D.M. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Chemmedchem / Year: 2021 Journal: Chemmedchem / Year: 2021Title: Structure-Based Drug Design of Phenazopyridine Derivatives as Inhibitors of Rev1 Interactions in Translesion Synthesis. Authors: McPherson, K.S. / Zaino, A.M. / Dash, R.C. / Rizzo, A.A. / Li, Y. / Hao, B. / Bezsonova, I. / Hadden, M.K. / Korzhnev, D.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ws5.cif.gz 6ws5.cif.gz | 136.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ws5.ent.gz pdb6ws5.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  6ws5.json.gz 6ws5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ws/6ws5 https://data.pdbj.org/pub/pdb/validation_reports/ws/6ws5 ftp://data.pdbj.org/pub/pdb/validation_reports/ws/6ws5 ftp://data.pdbj.org/pub/pdb/validation_reports/ws/6ws5 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6ws0C  3vu7S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 11011.666 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: REV1, REV1L / Production host: Homo sapiens (human) / Gene: REV1, REV1L / Production host:  References: UniProt: Q9UBZ9, Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases |
|---|---|
| #2: Protein | Mass: 26101.236 Da / Num. of mol.: 1 / Mutation: R124A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: MAD2L2, MAD2B, REV7 / Production host: Homo sapiens (human) / Gene: MAD2L2, MAD2B, REV7 / Production host:  |
| #3: Protein | Mass: 5632.319 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: REV3L, POLZ, REV3 / Production host: Homo sapiens (human) / Gene: REV3L, POLZ, REV3 / Production host:  |
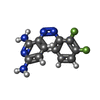
| #4: Chemical | ChemComp-U8M / |
| #5: Water | ChemComp-HOH / |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / Details: 162 mM triammonium citrate and 18% w/v PEG 3350 |
|---|
-Data collection
| Diffraction | Mean temperature: 280 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS-II NSLS-II  / Beamline: 17-ID-1 / Wavelength: 0.9201 Å / Beamline: 17-ID-1 / Wavelength: 0.9201 Å |
| Detector | Type: DECTRIS EIGER X 9M / Detector: PIXEL / Date: Sep 6, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9201 Å / Relative weight: 1 |
| Reflection | Resolution: 2.47→29.508 Å / Num. obs: 11712 / % possible obs: 99.6 % / Redundancy: 6.4 % / CC1/2: 0.999 / Rmerge(I) obs: 0.067 / Rpim(I) all: 0.028 / Rrim(I) all: 0.075 / Net I/σ(I): 13.7 |
| Reflection shell | Resolution: 2.47→2.57 Å / Rmerge(I) obs: 0.784 / Mean I/σ(I) obs: 2 / Num. unique obs: 1260 / CC1/2: 0.823 / Rpim(I) all: 0.47 / Rrim(I) all: 0.857 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3vu7 Resolution: 2.472→29.508 Å / Cor.coef. Fo:Fc: 0.962 / Cor.coef. Fo:Fc free: 0.948 / Cross valid method: FREE R-VALUE / ESU R: 0.913 / ESU R Free: 0.288 Details: Hydrogens have been added in their riding positions
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK BULK SOLVENT | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 70 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.472→29.508 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj