[English] 日本語
 Yorodumi
Yorodumi- PDB-6wmv: Structure of a phosphatidylinositol-phosphate synthase (PIPS) fro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6wmv | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii with evidence of substrate binding | ||||||||||||
 Components Components | AfCTD-Phosphatidylinositol-phosphate synthase (PIPS) fusion | ||||||||||||
 Keywords Keywords | MEMBRANE PROTEIN / CDP-alcohol phosphotransferase | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationTransferases; Transferring phosphorus-containing groups; Transferases for other substituted phosphate groups / phosphotransferase activity, for other substituted phosphate groups / phospholipid biosynthetic process / nucleotide binding / magnesium ion binding / metal ion binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Archaeoglobus fulgidus (archaea) Archaeoglobus fulgidus (archaea) Mycobacterium kansasii ATCC 12478 (bacteria) Mycobacterium kansasii ATCC 12478 (bacteria) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.142 Å MOLECULAR REPLACEMENT / Resolution: 2.142 Å | ||||||||||||
 Authors Authors | Belcher Dufrisne, M. / Jorge, C.D. / Timoteo, C.G. / Petrou, V.I. / Ashraf, K.U. / Banerjee, S. / Clarke, O.B. / Santos, H. / Mancia, F. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2020 Journal: J.Mol.Biol. / Year: 2020Title: Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria. Authors: Belcher Dufrisne, M. / Jorge, C.D. / Timoteo, C.G. / Petrou, V.I. / Ashraf, K.U. / Banerjee, S. / Clarke, O.B. / Santos, H. / Mancia, F. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6wmv.cif.gz 6wmv.cif.gz | 155.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6wmv.ent.gz pdb6wmv.ent.gz | 115.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6wmv.json.gz 6wmv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wm/6wmv https://data.pdbj.org/pub/pdb/validation_reports/wm/6wmv ftp://data.pdbj.org/pub/pdb/validation_reports/wm/6wmv ftp://data.pdbj.org/pub/pdb/validation_reports/wm/6wmv | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6wm5C  4o6mS  5d91S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Ens-ID: 1
|
- Components
Components
-Protein , 1 types, 2 molecules AC
| #1: Protein | Mass: 40388.797 Da / Num. of mol.: 2 / Mutation: D17L,Q77L,G79S Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Archaeoglobus fulgidus (archaea), (gene. exp.) Archaeoglobus fulgidus (archaea), (gene. exp.)  Mycobacterium kansasii ATCC 12478 (bacteria) Mycobacterium kansasii ATCC 12478 (bacteria)Gene: XD40_0003, XD48_0797, MKAN_26045 / Plasmid: pMCSG7 / Production host:  References: UniProt: A0A101DFK9, UniProt: U5WZP7, UniProt: O27985*PLUS |
|---|
-Non-polymers , 10 types, 53 molecules 

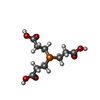




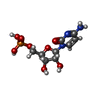











| #2: Chemical | ChemComp-LIP / | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-NA / #4: Chemical | ChemComp-TCE / | #5: Chemical | #6: Chemical | ChemComp-OLC / ( #7: Chemical | ChemComp-8K6 / #8: Chemical | #9: Chemical | ChemComp-C5P / | #10: Chemical | ChemComp-FLC / | #11: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.51 Å3/Da / Density % sol: 51.04 % |
|---|---|
| Crystal grow | Temperature: 295 K / Method: lipidic cubic phase / pH: 6.1 Details: 100 mM NaCl, 100 mM Sodium Citrate, pH 6.1, 40 mM MgCl2, 29% PEG 400, 500 uM L-myo-inositol-1-phosphate (prepared in house), 2.5 mM CDP (Sigma) (precipitant). Concentrated protein at 30-35 ...Details: 100 mM NaCl, 100 mM Sodium Citrate, pH 6.1, 40 mM MgCl2, 29% PEG 400, 500 uM L-myo-inositol-1-phosphate (prepared in house), 2.5 mM CDP (Sigma) (precipitant). Concentrated protein at 30-35 mg/ml was mixed with monoolein at a 1:1.5 protein to lipid ratio |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-E / Wavelength: 0.97918 Å / Beamline: 24-ID-E / Wavelength: 0.97918 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Mar 3, 2018 / Details: MD2 diffractometer |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97918 Å / Relative weight: 1 |
| Reflection | Resolution: 2.142→78.083 Å / Num. obs: 26006 / % possible obs: 58.72 % / Redundancy: 2 % / Biso Wilson estimate: 41.06 Å2 / CC1/2: 0.995 / CC star: 0.999 / Rmerge(I) obs: 0.07329 / Rpim(I) all: 0.07329 / Rrim(I) all: 0.1036 / Net I/σ(I): 9.78 |
| Reflection shell | Resolution: 2.142→2.221 Å / Redundancy: 2 % / Rmerge(I) obs: 0.4034 / Mean I/σ(I) obs: 1.73 / Num. unique obs: 69 / CC1/2: 0.594 / CC star: 0.863 / Rpim(I) all: 0.4034 / Rrim(I) all: 0.5705 / % possible all: 1.57 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5D91,4O6M Resolution: 2.142→78.083 Å / SU ML: 0.28 / Cross valid method: THROUGHOUT / σ(F): 1.37 / Phase error: 36.47
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.6 Å / VDW probe radii: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 116.15 Å2 / Biso mean: 45.381 Å2 / Biso min: 13.24 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.142→78.083 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0
|
 Movie
Movie Controller
Controller










 PDBj
PDBj



















