| 登録情報 | データベース: PDB / ID: 6w3t
|
|---|
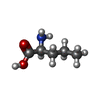
| タイトル | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-norvaline |
|---|
 要素 要素 | Methyl-accepting chemotaxis protein |
|---|
 キーワード キーワード | SIGNALING PROTEIN / Bacterial chemotaxis / chemoreceptor / double Cache / ligand binding domain |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
membrane => GO:0016020 / signal transduction / membrane類似検索 - 分子機能 Methyl-accepting chemotaxis protein-like, first PDC sensor domain / Double Cache domain 1 / Cache domain / Periplasmic sensor-like domain superfamily / Methyl-accepting chemotaxis protein (MCP) signalling domain / Methyl-accepting chemotaxis protein (MCP) signalling domain / Bacterial chemotaxis sensory transducers domain profile. / Methyl-accepting chemotaxis-like domains (chemotaxis sensory transducer).類似検索 - ドメイン・相同性 NORVALINE / Methyl-accepting chemotaxis protein / Methyl-accepting chemotaxis signal transduction protein類似検索 - 構成要素 |
|---|
| 生物種 |   Campylobacter jejuni (カンピロバクター) Campylobacter jejuni (カンピロバクター) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.1 Å 分子置換 / 解像度: 2.1 Å |
|---|
 データ登録者 データ登録者 | Khan, M.F. / Machuca, M.A. / Rahman, M.M. / Roujeinikova, A. |
|---|
| 資金援助 |  オーストラリア, 1件 オーストラリア, 1件 | 組織 | 認可番号 | 国 |
|---|
| Australian Research Council (ARC) | |  オーストラリア オーストラリア |
|
|---|
 引用 引用 |  ジャーナル: Biomolecules / 年: 2020 ジャーナル: Biomolecules / 年: 2020
タイトル: Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
著者: Khan, M.F. / Machuca, M.A. / Rahman, M.M. / Koc, C. / Norton, R.S. / Smith, B.J. / Roujeinikova, A. |
|---|
| 履歴 | | 登録 | 2020年3月9日 | 登録サイト: RCSB / 処理サイト: RCSB |
|---|
| 改定 1.0 | 2020年5月20日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2020年5月27日 | Group: Database references / カテゴリ: citation / citation_author
Item: _citation.journal_volume / _citation.pdbx_database_id_PubMed ..._citation.journal_volume / _citation.pdbx_database_id_PubMed / _citation.title / _citation_author.identifier_ORCID |
|---|
| 改定 1.2 | 2023年10月18日 | Group: Data collection / Database references / Refinement description
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| 改定 1.3 | 2024年10月9日 | Group: Structure summary
カテゴリ: pdbx_entry_details / pdbx_modification_feature
Item: _pdbx_entry_details.has_protein_modification |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.1 Å
分子置換 / 解像度: 2.1 Å  データ登録者
データ登録者 オーストラリア, 1件
オーストラリア, 1件  引用
引用 ジャーナル: Biomolecules / 年: 2020
ジャーナル: Biomolecules / 年: 2020 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 6w3t.cif.gz
6w3t.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb6w3t.ent.gz
pdb6w3t.ent.gz PDB形式
PDB形式 6w3t.json.gz
6w3t.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード https://data.pdbj.org/pub/pdb/validation_reports/w3/6w3t
https://data.pdbj.org/pub/pdb/validation_reports/w3/6w3t ftp://data.pdbj.org/pub/pdb/validation_reports/w3/6w3t
ftp://data.pdbj.org/pub/pdb/validation_reports/w3/6w3t リンク
リンク 集合体
集合体


 要素
要素

 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  Australian Synchrotron
Australian Synchrotron  / ビームライン: MX1 / 波長: 0.9537 Å
/ ビームライン: MX1 / 波長: 0.9537 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー





















 PDBj
PDBj