+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6u2v | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | AAV8 Baculovirus-Sf9 produced, full capsid | |||||||||||||||||||||||||||
 Components Components | Capsid protein | |||||||||||||||||||||||||||
 Keywords Keywords | VIRUS / Adeno-associated virus / AAV / gene therapy vector / post translational modification / capsid | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationT=1 icosahedral viral capsid / nucleotide binding / structural molecule activity Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Adeno-associated virus - 8 Adeno-associated virus - 8 | |||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||||||||||||||||||||
 Authors Authors | Paulk, N.K. / Poweleit, N. | |||||||||||||||||||||||||||
| Funding support |  United States, 8items United States, 8items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Mol Ther Methods Clin Dev / Year: 2020 Journal: Mol Ther Methods Clin Dev / Year: 2020Title: Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors. Authors: Neil G Rumachik / Stacy A Malaker / Nicole Poweleit / Lucy H Maynard / Christopher M Adams / Ryan D Leib / Giana Cirolia / Dennis Thomas / Susan Stamnes / Kathleen Holt / Patrick Sinn / ...Authors: Neil G Rumachik / Stacy A Malaker / Nicole Poweleit / Lucy H Maynard / Christopher M Adams / Ryan D Leib / Giana Cirolia / Dennis Thomas / Susan Stamnes / Kathleen Holt / Patrick Sinn / Andrew P May / Nicole K Paulk /  Abstract: Different approaches are used in the production of recombinant adeno-associated virus (rAAV). The two leading approaches are transiently transfected human HEK293 cells and live baculovirus infection ...Different approaches are used in the production of recombinant adeno-associated virus (rAAV). The two leading approaches are transiently transfected human HEK293 cells and live baculovirus infection of () insect cells. Unexplained differences in vector performance have been seen clinically and preclinically. Thus, we performed a controlled comparative production analysis varying only the host cell species but maintaining all other parameters. We characterized differences with multiple analytical approaches: proteomic profiling by mass spectrometry, isoelectric focusing, cryo-EM (transmission electron cryomicroscopy), denaturation assays, genomic and epigenomic sequencing of packaged genomes, human cytokine profiling, and functional transduction assessments and , including in humanized liver mice. Using these approaches, we have made two major discoveries: (1) rAAV capsids have post-translational modifications (PTMs), including glycosylation, acetylation, phosphorylation, and methylation, and these differ between platforms; and (2) rAAV genomes are methylated during production, and these are also differentially deposited between platforms. Our data show that host cell protein impurities differ between platforms and can have their own PTMs, including potentially immunogenic N-linked glycans. Human-produced rAAVs are more potent than baculovirus- vectors in various cell types (p < 0.05-0.0001), in various mouse tissues (p < 0.03-0.0001), and in human liver (p < 0.005). These differences may have clinical implications for rAAV receptor binding, trafficking, expression kinetics, expression durability, vector immunogenicity, as well as cost considerations. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6u2v.cif.gz 6u2v.cif.gz | 107.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6u2v.ent.gz pdb6u2v.ent.gz | 81.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6u2v.json.gz 6u2v.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  6u2v_validation.pdf.gz 6u2v_validation.pdf.gz | 1.3 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  6u2v_full_validation.pdf.gz 6u2v_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  6u2v_validation.xml.gz 6u2v_validation.xml.gz | 41.6 KB | Display | |
| Data in CIF |  6u2v_validation.cif.gz 6u2v_validation.cif.gz | 58.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/u2/6u2v https://data.pdbj.org/pub/pdb/validation_reports/u2/6u2v ftp://data.pdbj.org/pub/pdb/validation_reports/u2/6u2v ftp://data.pdbj.org/pub/pdb/validation_reports/u2/6u2v | HTTPS FTP |
-Related structure data
| Related structure data |  20626MC  6pwaC  6u20C  6ubmC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 | x 60
|
| 2 |
|
| 3 | x 5
|
| 4 | x 6
|
| 5 | 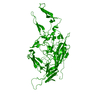
|
| Symmetry | Point symmetry: (Schoenflies symbol: I (icosahedral)) |
- Components
Components
| #1: Protein | Mass: 58528.367 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Adeno-associated virus - 8 / Production host: Adeno-associated virus - 8 / Production host:  |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Adeno-associated virus - 8 / Type: VIRUS Details: Recombinant AAV8 vector, produced in suspension Spodoptera frugiperda (Sf9) cells infected with live baculovirus expressing AAV components. Entity ID: all / Source: RECOMBINANT | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:  Adeno-associated virus - 8 Adeno-associated virus - 8 | ||||||||||||||||||||||||||||||
| Source (recombinant) | Organism:  | ||||||||||||||||||||||||||||||
| Details of virus | Empty: NO / Enveloped: NO / Isolate: SEROTYPE / Type: VIRION | ||||||||||||||||||||||||||||||
| Buffer solution | pH: 7.4 | ||||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 6 sec. / Electron dose: 66 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Num. of real images: 395 |
| Image scans | Movie frames/image: 30 |
- Processing
Processing
| EM software |
| |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||||||||||||||||||||
| Symmetry | Point symmetry: I (icosahedral) | |||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 404 / Symmetry type: POINT | |||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL |
 Movie
Movie Controller
Controller
















 PDBj
PDBj


