+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20502 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | AAV8 human HEK293-produced, full capsid | |||||||||||||||||||||||||||
 Map data Map data | AAV8 human HEK293-produced, full capsid | |||||||||||||||||||||||||||
 Sample Sample | Adeno-associated virus - 8 != Adeno-associated virus-8 Adeno-associated virus - 8
| |||||||||||||||||||||||||||
 Keywords Keywords | Adeno-associated virus / AAV / gene therapy vector / post translational modification / capsid / VIRUS | |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationT=1 icosahedral viral capsid / nucleotide binding / structural molecule activity Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Adeno-associated virus - 8 / Adeno-associated virus - 8 /  Adeno-associated virus-8 Adeno-associated virus-8 | |||||||||||||||||||||||||||
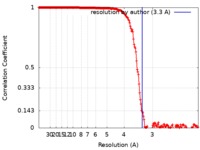
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||||||||||||||
 Authors Authors | Paulk NK / Poweleit N | |||||||||||||||||||||||||||
| Funding support |  United States, 8 items United States, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Mol Ther Methods Clin Dev / Year: 2020 Journal: Mol Ther Methods Clin Dev / Year: 2020Title: Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors. Authors: Neil G Rumachik / Stacy A Malaker / Nicole Poweleit / Lucy H Maynard / Christopher M Adams / Ryan D Leib / Giana Cirolia / Dennis Thomas / Susan Stamnes / Kathleen Holt / Patrick Sinn / ...Authors: Neil G Rumachik / Stacy A Malaker / Nicole Poweleit / Lucy H Maynard / Christopher M Adams / Ryan D Leib / Giana Cirolia / Dennis Thomas / Susan Stamnes / Kathleen Holt / Patrick Sinn / Andrew P May / Nicole K Paulk /  Abstract: Different approaches are used in the production of recombinant adeno-associated virus (rAAV). The two leading approaches are transiently transfected human HEK293 cells and live baculovirus infection ...Different approaches are used in the production of recombinant adeno-associated virus (rAAV). The two leading approaches are transiently transfected human HEK293 cells and live baculovirus infection of () insect cells. Unexplained differences in vector performance have been seen clinically and preclinically. Thus, we performed a controlled comparative production analysis varying only the host cell species but maintaining all other parameters. We characterized differences with multiple analytical approaches: proteomic profiling by mass spectrometry, isoelectric focusing, cryo-EM (transmission electron cryomicroscopy), denaturation assays, genomic and epigenomic sequencing of packaged genomes, human cytokine profiling, and functional transduction assessments and , including in humanized liver mice. Using these approaches, we have made two major discoveries: (1) rAAV capsids have post-translational modifications (PTMs), including glycosylation, acetylation, phosphorylation, and methylation, and these differ between platforms; and (2) rAAV genomes are methylated during production, and these are also differentially deposited between platforms. Our data show that host cell protein impurities differ between platforms and can have their own PTMs, including potentially immunogenic N-linked glycans. Human-produced rAAVs are more potent than baculovirus- vectors in various cell types (p < 0.05-0.0001), in various mouse tissues (p < 0.03-0.0001), and in human liver (p < 0.005). These differences may have clinical implications for rAAV receptor binding, trafficking, expression kinetics, expression durability, vector immunogenicity, as well as cost considerations. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20502.map.gz emd_20502.map.gz | 476.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20502-v30.xml emd-20502-v30.xml emd-20502.xml emd-20502.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20502_fsc.xml emd_20502_fsc.xml | 21.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_20502.png emd_20502.png | 115.2 KB | ||
| Filedesc metadata |  emd-20502.cif.gz emd-20502.cif.gz | 6.2 KB | ||
| Others |  emd_20502_half_map_1.map.gz emd_20502_half_map_1.map.gz emd_20502_half_map_2.map.gz emd_20502_half_map_2.map.gz | 46.4 MB 46.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20502 http://ftp.pdbj.org/pub/emdb/structures/EMD-20502 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20502 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20502 | HTTPS FTP |
-Related structure data
| Related structure data |  6pwaMC  6u20C  6u2vC  6ubmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20502.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20502.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AAV8 human HEK293-produced, full capsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
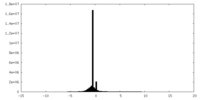
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: AAV8 human HEK293-produced, full capsid
| File | emd_20502_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AAV8 human HEK293-produced, full capsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
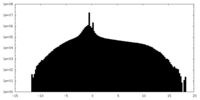
| Density Histograms |
-Half map: AAV8 human HEK293-produced, full capsid
| File | emd_20502_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | AAV8 human HEK293-produced, full capsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
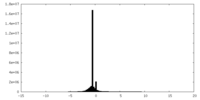
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus - 8
| Entire | Name:  Adeno-associated virus - 8 Adeno-associated virus - 8 |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus-8
| Supramolecule | Name: Adeno-associated virus-8 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Recombinant AAV8 vector, produced in adherent human HEK293 cells purchased from ATCC. NCBI-ID: 202813 / Sci species name: Adeno-associated virus-8 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Adeno-associated virus - 8 Adeno-associated virus - 8 |
| Molecular weight | Theoretical: 58.528367 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DGVGSSSGNW HCDSTWLGDR VITTSTRTWA LPTYNNHLYK QISNGTSGGA TNDNTYFGYS TPWGYFDFNR FHCHFSPRDW QRLINNNWG FRPKRLSFKL FNIQVKEVTQ NEGTKTIANN LTSTIQVFTD SEYQLPYVLG SAHQGCLPPF PADVFMIPQY G YLTLNNGS ...String: DGVGSSSGNW HCDSTWLGDR VITTSTRTWA LPTYNNHLYK QISNGTSGGA TNDNTYFGYS TPWGYFDFNR FHCHFSPRDW QRLINNNWG FRPKRLSFKL FNIQVKEVTQ NEGTKTIANN LTSTIQVFTD SEYQLPYVLG SAHQGCLPPF PADVFMIPQY G YLTLNNGS QAVGRSSFYC LEYFPSQMLR TGNNFQFTYT FEDVPFHSSY AHSQSLDRLM NPLIDQYLYY LSRTQTTGGT AN TQTLGFS QGGPNTMANQ AKNWLPGPCY RQQRVSTTTG QNNNSNFAWT AGTKYHLNGR NSLANPGIAM ATHKDDEERF FPS NGILIF GKQNAARDNA DYSDVMLTSE EEIKTTNPVA TEEYGIVADN LQQQNTAPQI GTVNSQGALP GMVWQNRDVY LQGP IWAKI PHTDGNFHPS PLMGGFGLKH PPPQILIKNT PVPADPPTTF NQSKLNSFIT QYSTGQVSVE IEWELQKENS KRWNP EIQY TSNYYKSTSV DFAVNTEGVY SEPRPIGTRY LTRNL UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Details: unspecified | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 1647 / Average exposure time: 6.0 sec. / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6pwa: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)