+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21656 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
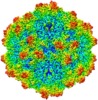
| Title | BatAAV-10HB - genome-containing particles | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Icosahedral Capsid / AAV / Gene Therapy / non-primate / Bat / VIRUS | |||||||||
| Function / homology | Phospholipase A2-like domain / Phospholipase A2-like domain / Parvovirus coat protein VP2 / Parvovirus coat protein VP1/VP2 / Parvovirus coat protein VP1/VP2 / Capsid/spike protein, ssDNA virus / T=1 icosahedral viral capsid / structural molecule activity / VP1 capsid Function and homology information Function and homology information | |||||||||
| Biological species |  Bat adeno-associated virus / Bat adeno-associated virus /   Adeno-associated virus Adeno-associated virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | |||||||||
 Authors Authors | Mietzsch M / Agbandje-McKenna M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2020 Journal: J Struct Biol / Year: 2020Title: Structural characterization of a bat Adeno-associated virus capsid. Authors: Mario Mietzsch / Ya Li / Justin Kurian / James Kennon Smith / Paul Chipman / Robert McKenna / Lin Yang / Mavis Agbandje-McKenna /   Abstract: Adeno-associated viruses (AAVs) are widespread among vertebrates. AAVs isolated from bats display low capsid protein sequence identities (<60%) to AAV2, AAV5, and other primate AAVs. Here we report the first capsid structure of a non-primate AAV which was isolated from bats. The capsid structure of BtAAV-10HB (10HB) was determined by cryo-electron microscopy and three-dimensional image reconstruction to 3.03 Å resolution. Comparison of empty and genome-containing capsids showed that the capsid structures are almost identical except for an ordered nucleotide in a previously described nucleotide-binding pocket, the density in the 5-fold channel, and several amino acids with altered side chain conformations. Compared to other dependoparvoviruses, for example AAV2 and AAV5, 10HB displays unique structural features including insertions and deletions in capsid surface loops. Overall, the 10HB capsid structure superposes with an RMSD of 1.7 Å and 1.8 Å to AAV2 and AAV5, respectively. Currently all approved AAV human gene therapy biologics and vectors in clinical trials are based on primate isolates. However, pre-existing neutralizing antibodies in the human population represents a hurdle to their use. 10HB capsids are capable of packaging AAV2 vector genomes and thus have potential as gene delivery vectors. Significantly, a screen with human sera showed lack of recognition by the 10HB capsid. Thus, the different capsid surface of 10HB vectors likely renders it "invisible" to potential pre-existing neutralizing human anti-AAV antibodies especially because this virus or similar variants do not exist in primate populations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21656.map.gz emd_21656.map.gz | 226.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21656-v30.xml emd-21656-v30.xml emd-21656.xml emd-21656.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21656.png emd_21656.png | 299.4 KB | ||
| Filedesc metadata |  emd-21656.cif.gz emd-21656.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21656 http://ftp.pdbj.org/pub/emdb/structures/EMD-21656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21656 | HTTPS FTP |
-Related structure data
| Related structure data |  6wftMC  6wfuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21656.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21656.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:   Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus
| Supramolecule | Name: Adeno-associated virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 272636 / Sci species name: Adeno-associated virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: VP1 capsid
| Macromolecule | Name: VP1 capsid / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bat adeno-associated virus Bat adeno-associated virus |
| Molecular weight | Theoretical: 57.741039 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DGVGQSSGNW HCDSVWMGDR VLTKSTRTWS LPTYNNHLYK QINGSGTGDA VYFGYSTPWG YFDFNRFHCH FSPRDWQRLV NNHWGIRPR RLNFKLFNIQ VKEVTTTDGT KTIANNLTST VQVFADTEHQ LPYILGSAHE GCMPPFPADV FMLPQYGYLT L NGPGSNNN ...String: DGVGQSSGNW HCDSVWMGDR VLTKSTRTWS LPTYNNHLYK QINGSGTGDA VYFGYSTPWG YFDFNRFHCH FSPRDWQRLV NNHWGIRPR RLNFKLFNIQ VKEVTTTDGT KTIANNLTST VQVFADTEHQ LPYILGSAHE GCMPPFPADV FMLPQYGYLT L NGPGSNNN NLSTPSSAFY CLEYFPSQML RTGNNFVFTY EFEKVPFHSM FMHNQALDRL MNPLVDQYLW YLDATSGNNL TF RKAGAKN FPEYFRNWIP GPGCRNQQWN KVGTKNNPQT GTWASANKWR LQGRLNKYAP GQPNAPAEGF LTNAGDLAFA NAK ATGATT AAGTVPADIL LTSESETTTT NMMSNNGWGA IASNNQNASV APTVQYEDSA HVLPGMVWQD RDIYLQGPIW AKIP ETDGH FHPSPLMGGF GLKNPPPQIL IKNTPVPADP PTQFSSQKIN SFITQYSTGQ MTVEIEWELR KENSKRWNPE IQYTA NFNN SANAQFSVNN NGLYIEDRTI GTRYLTHTL UniProtKB: VP1 capsid |
-Macromolecule #2: 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE
| Macromolecule | Name: 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE / type: ligand / ID: 2 / Number of copies: 60 / Formula: D5M |
|---|---|
| Molecular weight | Theoretical: 331.222 Da |
| Chemical component information | 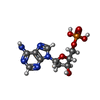 ChemComp-D5M: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)





















