| Entry | Database: PDB / ID: 6td2
|
|---|
| Title | Mus musculus Acetylcholinesterase in complex with N-(2-(diethylamino)ethyl)-1-(4-(trifluoromethyl)phenyl)methanesulfonamide |
|---|
 Components Components | Acetylcholinesterase |
|---|
 Keywords Keywords | HYDROLASE / Complex / Inhibitor |
|---|
| Function / homology |  Function and homology information Function and homology information
serine hydrolase activity / acetylcholine catabolic process / acetylcholinesterase / acetylcholine binding / acetylcholine receptor signaling pathway / osteoblast development / acetylcholinesterase activity / basement membrane / regulation of receptor recycling / side of membrane ...serine hydrolase activity / acetylcholine catabolic process / acetylcholinesterase / acetylcholine binding / acetylcholine receptor signaling pathway / osteoblast development / acetylcholinesterase activity / basement membrane / regulation of receptor recycling / side of membrane / laminin binding / collagen binding / neuromuscular junction / receptor internalization / positive regulation of cold-induced thermogenesis / retina development in camera-type eye / cell adhesion / synapse / perinuclear region of cytoplasm / cell surface / Golgi apparatus / protein homodimerization activity / extracellular space / identical protein binding / plasma membraneSimilarity search - Function Acetylcholinesterase, tetramerisation domain / Acetylcholinesterase tetramerisation domain / : / Cholinesterase / Carboxylesterase type B, active site / Carboxylesterases type-B serine active site. / Carboxylesterase type B, conserved site / Carboxylesterases type-B signature 2. / Carboxylesterase, type B / Carboxylesterase family / Alpha/Beta hydrolase foldSimilarity search - Domain/homology |
|---|
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 2.8 Å FOURIER SYNTHESIS / Resolution: 2.8 Å |
|---|
 Authors Authors | Forsgren, N. / Ekstrom, F. |
|---|
| Funding support | 2items | Organization | Grant number | Country |
|---|
| Other government | | | | Swedish Research Council | | |
|
|---|
 Citation Citation |  Journal: J.Phys.Chem.B / Year: 2020 Journal: J.Phys.Chem.B / Year: 2020
Title: Physical Mechanisms Governing Substituent Effects on Arene-Arene Interactions in a Protein Milieu.
Authors: Andersson, C.D. / Mishra, B.K. / Forsgren, N. / Ekstrom, F. / Linusson, A. |
|---|
| History | | Deposition | Nov 7, 2019 | Deposition site: PDBE / Processing site: PDBE |
|---|
| Revision 1.0 | Oct 14, 2020 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Oct 21, 2020 | Group: Derived calculations / Category: pdbx_struct_assembly / pdbx_struct_assembly_gen |
|---|
| Revision 1.2 | Jan 24, 2024 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
| Revision 1.3 | Oct 23, 2024 | Group: Structure summary / Category: pdbx_entry_details / pdbx_modification_feature / Item: _pdbx_entry_details.has_protein_modification |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 2.8 Å
FOURIER SYNTHESIS / Resolution: 2.8 Å  Authors
Authors Citation
Citation Journal: J.Phys.Chem.B / Year: 2020
Journal: J.Phys.Chem.B / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6td2.cif.gz
6td2.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6td2.ent.gz
pdb6td2.ent.gz PDB format
PDB format 6td2.json.gz
6td2.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/td/6td2
https://data.pdbj.org/pub/pdb/validation_reports/td/6td2 ftp://data.pdbj.org/pub/pdb/validation_reports/td/6td2
ftp://data.pdbj.org/pub/pdb/validation_reports/td/6td2
 Links
Links Assembly
Assembly

 Components
Components


 Homo sapiens (human) / References: UniProt: P21836, acetylcholinesterase
Homo sapiens (human) / References: UniProt: P21836, acetylcholinesterase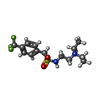









 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  MAX II
MAX II  / Beamline: I911-5 / Wavelength: 1.041 Å
/ Beamline: I911-5 / Wavelength: 1.041 Å Processing
Processing FOURIER SYNTHESIS
FOURIER SYNTHESIS Movie
Movie Controller
Controller






















 PDBj
PDBj





