[English] 日本語
 Yorodumi
Yorodumi- PDB-6ste: Crystal structure of the tick chemokine-binding protein Evasin-4 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ste | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the tick chemokine-binding protein Evasin-4 (SG 3) | ||||||
 Components Components | Evasin-4 | ||||||
 Keywords Keywords | IMMUNE SYSTEM / CHEMOKINE-BINDING PROTEIN / TICKS | ||||||
| Function / homology | negative regulation of chemokine activity / Evasins Class A / Evasins Class A / C-C chemokine binding / Immunoglobulin-like domain / extracellular region / ACETATE ION / DI(HYDROXYETHYL)ETHER / Evasin-4 Function and homology information Function and homology information | ||||||
| Biological species |  Rhipicephalus sanguineus (brown dog tick) Rhipicephalus sanguineus (brown dog tick) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.79 Å MOLECULAR REPLACEMENT / Resolution: 1.79 Å | ||||||
 Authors Authors | Ramirez-Escudero, M. / Janssen, B.J.C. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2020 Journal: J.Biol.Chem. / Year: 2020Title: Structural characterization of anti-CCL5 activity of the tick salivary protein evasin-4. Authors: Denisov, S.S. / Ramirez-Escudero, M. / Heinzmann, A.C.A. / Ippel, J.H. / Dawson, P.E. / Koenen, R.R. / Hackeng, T.M. / Janssen, B.J.C. / Dijkgraaf, I. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ste.cif.gz 6ste.cif.gz | 150.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ste.ent.gz pdb6ste.ent.gz | 118.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ste.json.gz 6ste.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/st/6ste https://data.pdbj.org/pub/pdb/validation_reports/st/6ste ftp://data.pdbj.org/pub/pdb/validation_reports/st/6ste ftp://data.pdbj.org/pub/pdb/validation_reports/st/6ste | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6st4SC 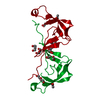 6stcC  6stkC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Ens-ID: 1
|
 Movie
Movie Controller
Controller










 PDBj
PDBj