[English] 日本語
 Yorodumi
Yorodumi- PDB-6prp: Structural Basis for Client Recognition and Activity of Hsp40 Cha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6prp | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structural Basis for Client Recognition and Activity of Hsp40 Chaperones | ||||||
 Components Components | Chaperone protein DnaK, Chaperone protein DnaJ 2 fusion | ||||||
 Keywords Keywords | CHAPERONE / HYDROLASE / Client Recognition | ||||||
| Function / homology |  Function and homology information Function and homology information: / ATP-dependent protein folding chaperone / unfolded protein binding / protein refolding / DNA replication / ATP binding / cytoplasm Similarity search - Function | ||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | ||||||
| Method | SOLUTION NMR / molecular dynamics | ||||||
 Authors Authors | Jiang, Y. / Rossi, P. / Kalodimos, C.G. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Structural basis for client recognition and activity of Hsp40 chaperones. Authors: Jiang, Y. / Rossi, P. / Kalodimos, C.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6prp.cif.gz 6prp.cif.gz | 579.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6prp.ent.gz pdb6prp.ent.gz | 494.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6prp.json.gz 6prp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pr/6prp https://data.pdbj.org/pub/pdb/validation_reports/pr/6prp ftp://data.pdbj.org/pub/pdb/validation_reports/pr/6prp ftp://data.pdbj.org/pub/pdb/validation_reports/pr/6prp | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6pptC  6pq2C  6pqeC  6pqmC  6priC  6prjC  6prqC  6psiC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 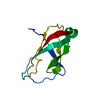
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 9176.394 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Thermus thermophilus (bacteria), (gene. exp.) Thermus thermophilus (bacteria), (gene. exp.)   Thermus thermophilus (strain HB8 / ATCC 27634 / DSM 579) (bacteria) Thermus thermophilus (strain HB8 / ATCC 27634 / DSM 579) (bacteria)Strain: HB8 / ATCC 27634 / DSM 579 / Gene: dnaJ2, TTHA1489 / Production host:  References: UniProt: Q56237, UniProt: Q56235*PLUS, alkaline phosphatase |
|---|
-Experimental details
-Experiment
| Experiment | Method: SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Type: solution Contents: 1 mM [U-100% 13C; U-100% 15N] DnaK-ctail-CBD2, 75 mM potassium chloride, 20 mM potassium phosphate, 0.04 % sodium azide, 90% H2O/10% D2O Details: double labeled fusion / Label: 15N_13C_sample / Solvent system: 90% H2O/10% D2O | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||
| Sample conditions | Ionic strength: 100 mM / Label: conditions_1 / pH: 7 / Pressure: 1 atm / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker AVANCE NEO / Manufacturer: Bruker / Model: AVANCE NEO / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: molecular dynamics / Software ordinal: 1 / Details: restrained MD in explicit H2O | |||||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | |||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: target function / Conformers calculated total number: 100 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller











 PDBj
PDBj


 HSQC
HSQC