[English] 日本語
 Yorodumi
Yorodumi- PDB-6pna: Structure of human neuronal nitric oxide synthase R354A/G357D mut... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6pna | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(4-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | ||||||
 Components Components | Nitric oxide synthase, brain | ||||||
 Keywords Keywords | OXIDOREDUCTASE/Inhibitor / nitric oxide synthase inhibitor / heme enzyme / OXIDOREDUCTASE / OXIDOREDUCTASE-Inhibitor complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of membrane repolarization during ventricular cardiac muscle cell action potential / negative regulation of calcium ion transport into cytosol / Nitric oxide stimulates guanylate cyclase / myoblast fusion / ROS and RNS production in phagocytes / tetrahydrobiopterin binding / arginine binding / regulation of cardiac muscle contraction by calcium ion signaling / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / synaptic signaling by nitric oxide ...positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential / negative regulation of calcium ion transport into cytosol / Nitric oxide stimulates guanylate cyclase / myoblast fusion / ROS and RNS production in phagocytes / tetrahydrobiopterin binding / arginine binding / regulation of cardiac muscle contraction by calcium ion signaling / regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / synaptic signaling by nitric oxide / positive regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / positive regulation of sodium ion transmembrane transport / peptidyl-cysteine S-nitrosylase activity / positive regulation of the force of heart contraction / cadmium ion binding / negative regulation of calcium ion transport / negative regulation of potassium ion transport / regulation of postsynaptic membrane potential / nitric oxide mediated signal transduction / nitric-oxide synthase (NADPH) / sodium channel regulator activity / regulation of neurogenesis / negative regulation of serotonin uptake / nitric-oxide synthase activity / regulation of cardiac muscle contraction / xenobiotic catabolic process / L-arginine catabolic process / multicellular organismal response to stress / striated muscle contraction / negative regulation of blood pressure / regulation of sodium ion transport / response to hormone / Ion homeostasis / nitric oxide biosynthetic process / photoreceptor inner segment / T-tubule / sarcoplasmic reticulum membrane / cell redox homeostasis / calyx of Held / cell periphery / sarcoplasmic reticulum / calcium channel regulator activity / establishment of localization in cell / potassium ion transport / sarcolemma / caveola / cellular response to growth factor stimulus / vasodilation / Z disc / calcium-dependent protein binding / calcium ion transport / FMN binding / NADP binding / flavin adenine dinucleotide binding / positive regulation of neuron apoptotic process / response to heat / scaffold protein binding / response to lipopolysaccharide / dendritic spine / transmembrane transporter binding / cytoskeleton / response to hypoxia / calmodulin binding / postsynaptic density / membrane raft / heme binding / synapse / positive regulation of DNA-templated transcription / perinuclear region of cytoplasm / positive regulation of transcription by RNA polymerase II / protein-containing complex / mitochondrion / nucleoplasm / nucleus / plasma membrane / cytoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 1.95 Å FOURIER SYNTHESIS / Resolution: 1.95 Å | ||||||
 Authors Authors | Li, H. / Poulos, T.L. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J.Med.Chem. / Year: 2020 Journal: J.Med.Chem. / Year: 2020Title: First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate. Authors: Cinelli, M.A. / Reidl, C.T. / Li, H. / Chreifi, G. / Poulos, T.L. / Silverman, R.B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6pna.cif.gz 6pna.cif.gz | 370.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6pna.ent.gz pdb6pna.ent.gz | 300.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6pna.json.gz 6pna.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pn/6pna https://data.pdbj.org/pub/pdb/validation_reports/pn/6pna ftp://data.pdbj.org/pub/pdb/validation_reports/pn/6pna ftp://data.pdbj.org/pub/pdb/validation_reports/pn/6pna | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6pmvC  6pmwC  6pmxC  6pmyC  6pmzC  6pn0C  6pn1C  6pn2C  6pn3C  6pn4C  6pn5C  6pn6C  6pn7C  6pn8C  6pn9C  6pnbC  6pncC  6pndC  6pneC  6pnfC  6pngC  6pnhC  6po5C  6po7C  6po8C  6po9C  6poaC  6pobC  6pocC  6potC  6pouC  6povC  6powC  6poxC  6poyC  6pozC  6pp0C  6pp1C  6pp2C  6pp3C  6pp4C  4uh5S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein | Mass: 48784.496 Da / Num. of mol.: 2 / Mutation: R354A, G357D Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: NOS1 / Organ: brain / Plasmid: pCWori / Production host: Homo sapiens (human) / Gene: NOS1 / Organ: brain / Plasmid: pCWori / Production host:  |
|---|
-Non-polymers , 6 types, 756 molecules 

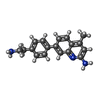








| #2: Chemical | | #3: Chemical | #4: Chemical | #5: Chemical | ChemComp-GOL / | #6: Chemical | ChemComp-ZN / | #7: Water | ChemComp-HOH / | |
|---|
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.74 Å3/Da / Density % sol: 55.2 % / Description: plates |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 7.2 Details: 8% PEG3350 35mM citric acid 65mM Bis-Tris-Propane 10% glycerol 5mM TCEP |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL14-1 / Wavelength: 0.9795 Å / Beamline: BL14-1 / Wavelength: 0.9795 Å |
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Jan 22, 2017 / Details: mirrors |
| Radiation | Monochromator: DOUBLE CRYSTAL SI(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 1.95→60 Å / Num. obs: 76617 / % possible obs: 99 % / Observed criterion σ(I): -3 / Redundancy: 4 % / CC1/2: 0.989 / Rmerge(I) obs: 0.141 / Rpim(I) all: 0.121 / Rsym value: 0.141 / Net I/σ(I): 5.3 |
| Reflection shell | Resolution: 1.95→2.01 Å / Redundancy: 3.9 % / Rmerge(I) obs: 2.087 / Mean I/σ(I) obs: 0.7 / Num. unique obs: 4364 / CC1/2: 0.315 / Rpim(I) all: 1.843 / Rsym value: 2.087 / % possible all: 97 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: 4UH5 Resolution: 1.95→43.993 Å / SU ML: 0.31 / Cross valid method: FREE R-VALUE / σ(F): 0.03 / Phase error: 29.17
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.95→43.993 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj










