[English] 日本語
 Yorodumi
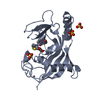
Yorodumi- PDB-6oxb: First bromo-adjacent homology (BAH) domain of human Polybromo-1 (... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6oxb | ||||||
|---|---|---|---|---|---|---|---|
| Title | First bromo-adjacent homology (BAH) domain of human Polybromo-1 (PBRM1) | ||||||
 Components Components | Protein polybromo-1 | ||||||
 Keywords Keywords | DNA BINDING PROTEIN / SWI/SNF chromatin remodeler / PBAF / bromodomain / BAH / acidic patch / nucleosome / DNA binding | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of G0 to G1 transition / regulation of nucleotide-excision repair / RSC-type complex / SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of T cell differentiation / positive regulation of double-strand break repair / nuclear chromosome / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / regulation of G1/S transition of mitotic cell cycle ...regulation of G0 to G1 transition / regulation of nucleotide-excision repair / RSC-type complex / SWI/SNF complex / regulation of mitotic metaphase/anaphase transition / positive regulation of T cell differentiation / positive regulation of double-strand break repair / nuclear chromosome / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / regulation of G1/S transition of mitotic cell cycle / positive regulation of myoblast differentiation / positive regulation of cell differentiation / transcription elongation by RNA polymerase II / kinetochore / RMTs methylate histone arginines / nuclear matrix / mitotic cell cycle / chromatin remodeling / negative regulation of cell population proliferation / chromatin binding / regulation of transcription by RNA polymerase II / chromatin / DNA binding / nucleoplasm / nucleus Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.86 Å molecular replacement / Resolution: 1.86 Å | ||||||
 Authors Authors | Petojevic, T. / Holliday, M.J. / Fairbrother, W.J. / Cochran, A.G. | ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Polyvalent nucleosome recognition by Polybromo-1 anchors chromatin remodeling Authors: Petojevic, T. / Holliday, M.J. / Dimitrova, Y.N. / Noland, C.L. / Fairbrother, W.J. / Cochran, A.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6oxb.cif.gz 6oxb.cif.gz | 430.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6oxb.ent.gz pdb6oxb.ent.gz | 353 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6oxb.json.gz 6oxb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ox/6oxb https://data.pdbj.org/pub/pdb/validation_reports/ox/6oxb ftp://data.pdbj.org/pub/pdb/validation_reports/ox/6oxb ftp://data.pdbj.org/pub/pdb/validation_reports/ox/6oxb | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1w4sS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Ens-ID: 1
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




