[English] 日本語
 Yorodumi
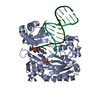
Yorodumi- PDB-6lgq: The crystal complex structure of histidine kinase and response re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6lgq | ||||||
|---|---|---|---|---|---|---|---|
| Title | The crystal complex structure of histidine kinase and response regulator | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SIGNALING PROTEIN / Two-component system / histidine kinase / response regulator | ||||||
| Function / homology |  Function and homology information Function and homology informationtransferase activity, transferring phosphorus-containing groups / cellular response to potassium ion / phosphorelay response regulator activity / DNA-binding transcription activator activity / phosphorelay sensor kinase activity / histidine kinase / phosphorelay signal transduction system / cis-regulatory region sequence-specific DNA binding / protein-DNA complex / outer membrane-bounded periplasmic space ...transferase activity, transferring phosphorus-containing groups / cellular response to potassium ion / phosphorelay response regulator activity / DNA-binding transcription activator activity / phosphorelay sensor kinase activity / histidine kinase / phosphorelay signal transduction system / cis-regulatory region sequence-specific DNA binding / protein-DNA complex / outer membrane-bounded periplasmic space / transcription cis-regulatory region binding / DNA-binding transcription factor activity / regulation of DNA-templated transcription / signal transduction / protein homodimerization activity / DNA binding / ATP binding / membrane / plasma membrane / cytosol Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3 Å MOLECULAR REPLACEMENT / Resolution: 3 Å | ||||||
 Authors Authors | Ming, Q.X. | ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The crystal complex structure of histidine kinase and response regulator Authors: Ming, Q.X. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6lgq.cif.gz 6lgq.cif.gz | 122.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6lgq.ent.gz pdb6lgq.ent.gz | 75.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6lgq.json.gz 6lgq.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/lg/6lgq https://data.pdbj.org/pub/pdb/validation_reports/lg/6lgq ftp://data.pdbj.org/pub/pdb/validation_reports/lg/6lgq ftp://data.pdbj.org/pub/pdb/validation_reports/lg/6lgq | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4knyS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 27321.896 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: kdbD, kdpD, AKG99_04800, AML35_19290, B9M99_01250, BK383_18195, C5P44_02760, CIJ94_01275, D2188_15845, D9H70_02790, D9J60_00745, DTL90_04085, DU321_01600, EC382_20775, EIA21_11550, EO241_08335, ...Gene: kdbD, kdpD, AKG99_04800, AML35_19290, B9M99_01250, BK383_18195, C5P44_02760, CIJ94_01275, D2188_15845, D9H70_02790, D9J60_00745, DTL90_04085, DU321_01600, EC382_20775, EIA21_11550, EO241_08335, EXX06_11290, EXX23_12490, EXX53_15910, EXX55_10750, EXX87_15770, FNJ83_05465, HMPREF3040_04620, NCTC9119_03628, SAMEA3472056_00110 Production host:  References: UniProt: A0A069C8B3, UniProt: P21865*PLUS, histidine kinase |
|---|---|
| #2: Protein | Mass: 27206.107 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: kdpE_1, kdpE, A8M42_04350, ACU57_12825, AM464_20745, AUQ13_21380, B9T59_04520, BANRA_01744, BANRA_03868, BEN53_05945, BK248_02745, BK334_19180, BON75_19570, BON86_22395, BvCmsHHP001_00776, ...Gene: kdpE_1, kdpE, A8M42_04350, ACU57_12825, AM464_20745, AUQ13_21380, B9T59_04520, BANRA_01744, BANRA_03868, BEN53_05945, BK248_02745, BK334_19180, BON75_19570, BON86_22395, BvCmsHHP001_00776, BvCmsHHP019_01442, BvCmsKSP026_01795, BvCmsSINP011_01149, BW690_07930, C4J69_07925, C4M78_23895, C5N07_14640, C6B13_12830, C9200_17795, CA593_25685, CDL37_26920, CI694_20635, CQP61_19525, CSB64_10560, D2183_02080, D2185_15895, D3821_21875, D3O91_17430, D9D20_12870, DAH34_18930, DB359_02985, DBQ99_18030, DM129_15030, DNQ41_07435, DNX30_24225, DTM25_00915, DWB25_17365, E2119_26455, E2134_18575, EAI52_22440, EC3234A_8c00210, EC3426_01519, ECONIH1_03925, ECTO6_03354, EIA08_00640, EJH97_16770, EL75_3094, EL79_3187, EL80_5454, ELT20_10810, ELT49_12200, ELT58_16530, EPS71_20965, EPS91_08680, EPT01_08635, ExPECSC022_01393, ExPECSC038_02992, EXX71_13740, EYD11_16000, FTV90_11805, FV293_12605, FV295_01400, MS6198_07470, NCTC10766_06081, NCTC13462_01629, NCTC13846_03346, NCTC7922_06598, NCTC7927_03942, NCTC7928_05334, NCTC8450_01686, NCTC9007_02376, NCTC9045_04052, NCTC9062_04757, NCTC9117_04356, NCTC9706_00828, NCTC9777_03550, RK56_025185, SAMEA3472033_02545, SAMEA3472047_02080, SAMEA3472080_00274, SAMEA3484427_03082, SAMEA3484429_03300, SAMEA3752559_01043, SAMEA3753300_04142, SK85_00691 Production host:  References: UniProt: A0A061KF22, UniProt: P21866*PLUS |
| #3: Water | ChemComp-HOH / |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.43 Å3/Da / Density % sol: 49.31 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / Details: 0.1M HEPES pH 7.0, 5% PEG6000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRF SSRF  / Beamline: BL17U / Wavelength: 0.97918 Å / Beamline: BL17U / Wavelength: 0.97918 Å |
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: Dec 25, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.97918 Å / Relative weight: 1 |
| Reflection | Resolution: 3→50 Å / Num. obs: 10464 / % possible obs: 92.21 % / Redundancy: 3.9 % / Biso Wilson estimate: 49.56 Å2 / Rpim(I) all: 0.026 / Net I/σ(I): 28.84 |
| Reflection shell | Resolution: 3→3.05 Å / Num. unique obs: 455 / CC1/2: 0.68 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4KNY Resolution: 3→38.83 Å / SU ML: 0.3651 / Cross valid method: FREE R-VALUE / σ(F): 1.39 / Phase error: 22.4876
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.06 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3→38.83 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj




